Danish researchers reveal how the MRSA bacterium handles stress
An international team of researchers has revealed a fundamental mechanism responsible for handling stress in staphylococci when they are exposed to antibiotics. It is expected that the research results eventually can be used to develop new antibiotics that circumvent such stress mechanisms.
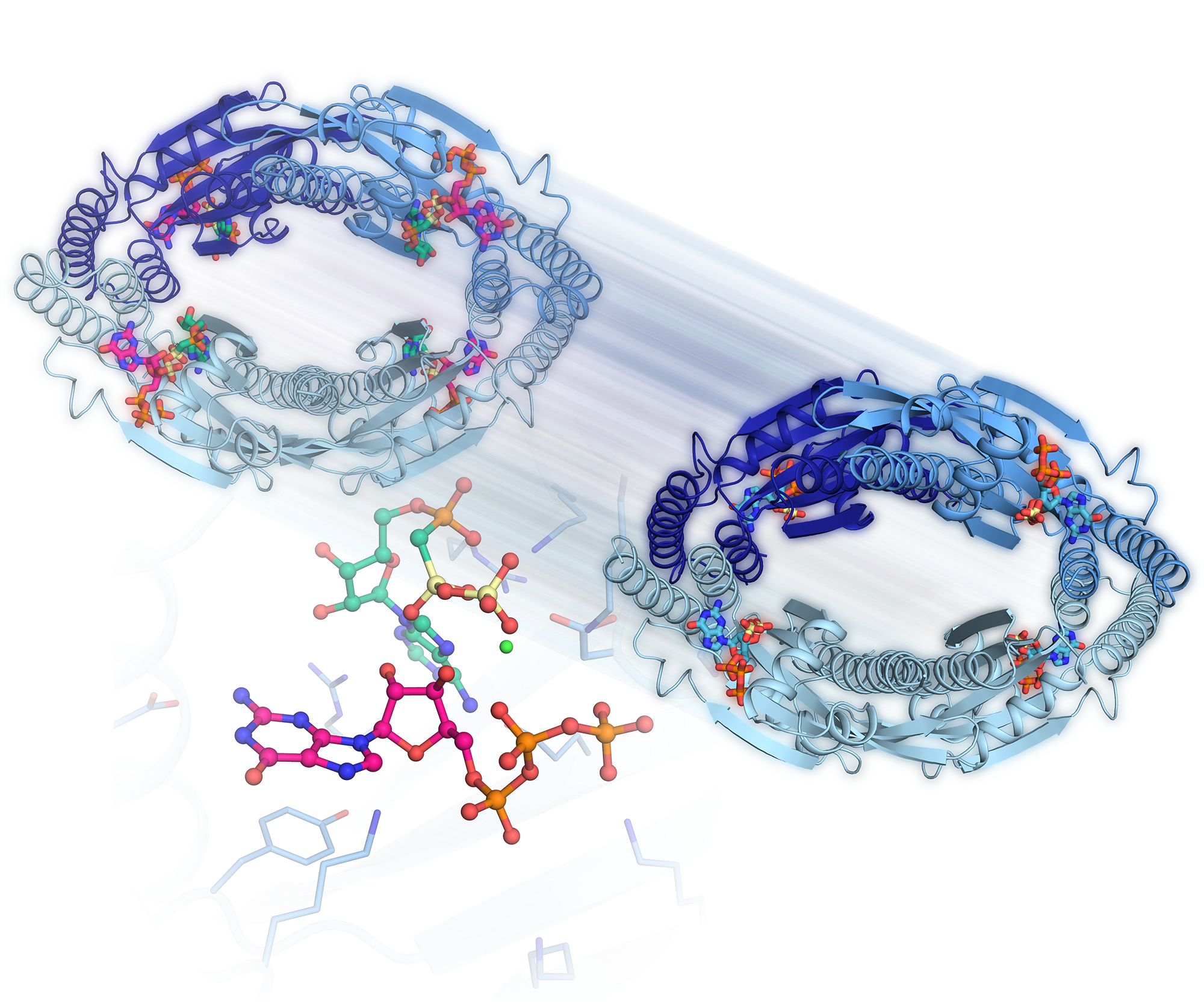
Understanding bacterial stress mechanisms is of great importance for our ability to treat bacterial infections, as these mechanisms often allow bacteria to survive antibiotic treatment. A group of researchers at the Department of Molecular Biology and Genetics at Aarhus University has now (in collaboration with researchers at the University of Copenhagen, Umeå University in Sweden, and Tartu University in Estonia) determined a fundamental mechanism by which the staphylococci bacteria, which are responsible for MRSA, handle stress when exposed to e.g. antibiotics.
The new research results, just published in the American Journal of Biological Chemistry, show how the bacteria are able to produce a particular enzyme capable of modifying some of the building blocks commonly used for the genetic material DNA, to turn them into signal molecules that signal stress. When the bacterial cells are exposed to e.g. antibiotics, large amounts of these signal molecules are formed resulting in a halt in cell growth and entry into a state of hibernation, where the cells are not so susceptible to antibiotics and are thus able to survive.
The researchers from Aarhus University, who have worked under the direction of Associate Professor Ditlev E. Brodersen, have used a refined experimental technique to establish accurate three-dimensional models for the states of the enzyme just before and just after it has formed a signal molecule. The models, which are accurate down to the atomic level, reveal how the enzyme participates in the reaction that leads to the formation of the signal molecule. At the same time, the researchers revealed that four enzymes unite to form a circle inside the bacterial cells, thus being able to communicate with each other. This feature may significantly increase the formation of signal substances during antibiotic treatment, thus increasing the overall effect. It is expected that the research results will eventually be used to develop new antibiotics that circumvent stress mechanisms.
Link to the article in Journal of Biological Chemistry med titlen "Structural basis for (p)ppGpp synthesis by the Staphylococcus aureus small alarmone synthetase RelP".
For further information, please contact
Associate Professor Ditlev E. Brodersen
Department of Molecular Biology and Genetics, Aarhus University, Denmark
deb@mbg.au.dk - +45 2166 9001
