New protein complexes shed light on quality control in human cells
Researchers at Aarhus University have identified and characterised two novel protein complexes associated with the human RNA exosome, an important player in the cell's quality control of RNA. The new protein complexes will help determine how RNA molecules are discriminated by the cell, and the discovery, which is published in the prestigious international journal Molecular Cell, is therefore an important step forward in understanding how human cells protect themselves against errors in gene expression.
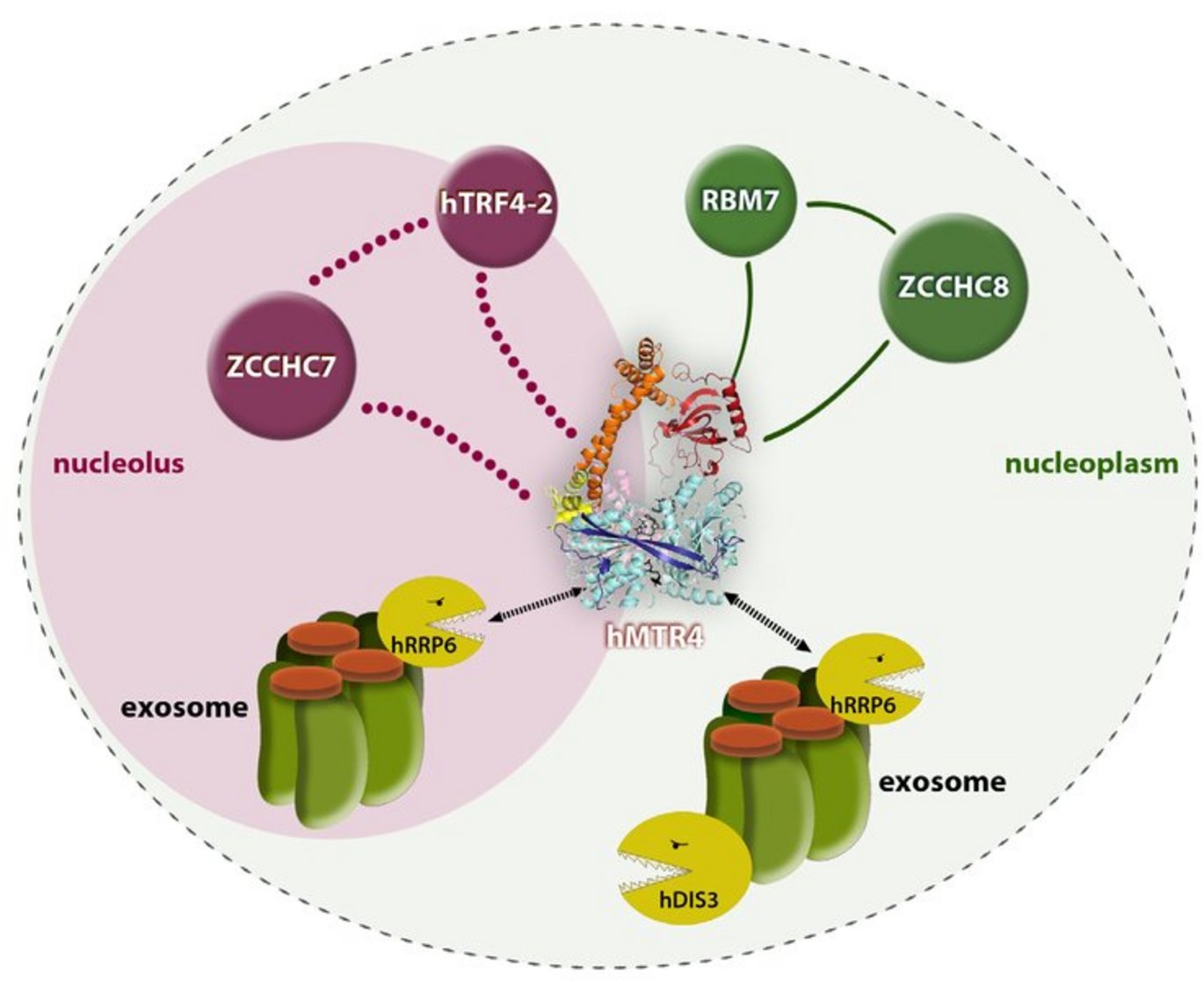
The exosome performs quality control
The RNA exosome is a complex machine consisting of nine core proteins that form a catalytically inactive ring-shaped structure onto which the active components are positioned. It is involved in the processing and degradation of RNA molecules that are essential for virtually all cellular processes. Besides removing RNAs that are no longer functional, the exosome performs quality control, which ensures that defective molecules do not create hazardous conditions inside the cell. Therefore, it is not surprising that the RNA exosome is evolutionarily conserved and can also be found in yeast, flies and plants.
A major scientific issue has been to clarify how the exosome identifies the substrates to be removed and which ones to leave in peace. Previous results from the centre have shown the existence of three different catalytic and exosome-associated proteins in humans, and that these are located differently inside the cell (see media release of 31 August 2010). While this spatial separation is part of the explanation of how exosome substrates are chosen, it is not sufficient to explain the substrate specificity – i.e. which RNAs are recognised.
The discovery of two novel protein complexes
In yeast, the exosome is helped by a protein complex called TRAMP (Trf4p/Air1p/Mtr4p polyadenylation), which contains an RNA-binding protein (Air1p) that directs the RNA to the exosome. In addition, a helicase (Mtr4p) eliminates internal constraints in the RNA molecule, and a polyA polymerase (Trf4p) extends the RNA hereby creating a handle for the exosome if it gets stuck or falls off. A similar auxiliary complex has not previously been described in human cells.
The present study led to the discovery of no fewer than two auxiliary complexes in human cells. One – coined human TRAMP (hTRAMP) – consists of proteins that correspond evolutionarily to the complex in yeast (hTrf4-2/ZCCHC7/hMtr4), while another – coined the Nuclear EXosome Targeting (NEXT) complex – contains two potential RNA-binding proteins (RBM7 and ZCCHC8) and the helicase hMtr4, which thus appears in both complexes.
A characterisation of the complexes demonstrated that the NEXT complex is extremely stable and can be found in most of the cell nucleus, except in specialised areas called nucleoli. In contrast, the hTRAMP appeared to be considerably less stable and was found exclusively in nucleoli.
This spatial separation was also reflected by the substrate specificity of the two complexes; NEXT is essential for the degradation of a heterogeneous class of unstable RNAs, called PROMPTs, which are degraded rapidly in the nucleoplasm, while hTRAMP degrades defective RNAs that accumulate in nucleoli.
Future studies will aim to identify yet additional RNA substrates for these new ‘exosome adapters’.
These studies were conducted at the Danish National Research Foundation Centre for mRNP Biogenesis and Metabolism located at the Department of Molecular Biology and Genetics at Aarhus University. The work was a collaboration between the three graduate students Michal Lubas, Marianne S. Christensen and Maiken S. Kristiansen from Professor Torben Heick Jensen's research group. The research groups of Andrzej Dziembowski, Warsaw University and Jens S. Andersen, University of Southern Denmark were involved as collaborators.
Link to the article:
Interaction Profiling Identifies the Human Nuclear EXosome Targeting (NEXT) Complex
Further information
 | Professor Torben Heick Jensen Centre for mRNP Biogenesis and Metabolism Department of Molecular Biology and Genetics, Aarhus University, Denmark Cell: +45 6020 2705, thj@mb.au.dk, www.mRNP.dk | |||
Related news:
- Internal communication within genes stimulates gene expression (12.11.2010)
- New catalytic subunit of the human RNA exosome (31.08.2010)
- Danish-Swiss research collaboration sheds new light on vital cellular process (08.12.2008)
- Revelation of previously hidden transcription activities in the human genome (05.12.2008)
- Molecular mechanism behind quality control of gene expression revealed (10.07.2008)
Text: Michal Lubas, Torben Heick Jensen and Lisbeth Heilesen
Translation: Lisbeth Heilesen
