Scientists reveal new basic knowledge of cell membranes
Following an interdisciplinary collaboration, scientists at Aarhus University have presented a ground-breaking method to study cell membranes. A better understanding of the complexity of membrane function may give the scientists new ideas for developing new types of medical treatment against prostate cancer, for example. The results have been published in the leading journal Nature Communications.

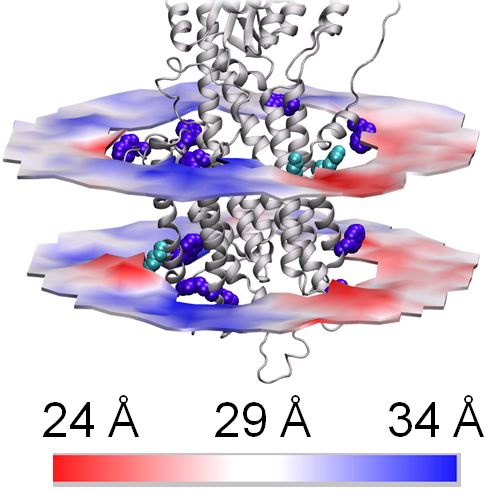
By combining many disciplines from the Danish National Research Foundation Centres PUMPKIN and inSPIN, scientists have shed new light on the structure and function of cell membranes based on computer calculations supported by experimental structural studies.
Cells and cell membranes have been known ever since the microscope was invented, but scientists now seek to link general observations on the physiology of cells and organisms, as well as health and illness, to details of molecular structure and function to understand the fundamental processes that define life and interaction between genes and the environment.
Membranes surround the cells and their room divisions and are therefore essential for all life. They consist of fats and proteins and are the target of approximately half of our known drugs. The complex interactions between proteins and fats in the membrane are critical for membrane function, but are very difficult to study by experiments alone. However, the Aarhus scientists have now succeeded in this by combining computer-based calculations, which has opened a door to new insights into molecular cell biology.
An innovative combination of X-ray crystallographic data collected on X-ray synchrotron sources and molecular dynamic simulations carried out on powerful computers gave the research team a unique basis on which to visualise and study the interaction between the membrane and the “calcium pump” – a membrane protein that pumps calcium across the cell membrane during major structural changes. An essential factor was being able to show that the experiments and the independently established computer models gave consistent results. The study therefore enables a clarification of questions about how membrane proteins are embedded in cell membranes and can also be used to study the effect of drugs on membrane proteins.
“We discovered all in one go that we could explore how an interesting membrane protein is embedded in the cell membranes, and how it changes when it works,” explains Associate Professor Lea Thøgersen, who was in charge of the computer calculations in the study along with PhD Student Maria Musgaard. “Compared with the past, where we had only schematic models, we can now describe the protein's interaction with the membrane at a molecular level that helps us explain the function of a key protein,” adds Maria Musgaard. “This is essential for understanding how the transport functions of the cells change in connection with illness and drug administration,” she continues.
A greater understanding of this complexity in the function of the membrane proteins may give the scientists new ideas for developing new types of medical treatment against prostate cancer, for example. Here the focus is on the calcium transport of cancer cells – because if this is blocked, the cancer cells die.
Professor Jesper Vuust Møller, who has been involved with calcium transport for many years, says on this occasion, “It has always been my dream as a researcher to get new ideas and insight into nature and, as a medical scientist, to find at the same time the way to new methods of treatment against major diseases such as prostate cancer.” The Danish National Research Foundation Centre PUMPKIN studies prostate cancer research in collaboration with scientists in Copenhagen, USA and France.
“It is the combination of many disciplines that has been crucial to this success story,” says PUMPKIN Centre Director Professor Poul Nissen. “We’ve brought physiology, biophysics, molecular biology and chemistry together in this project – it’s in cross fields like these that exciting new questions and insights arise, both now and in the future,” he adds.
The project’s interdisciplinary character is reflected in the composition of the research team. The authors are affiliated with the Danish National Research Foundation Centres PUMPKIN and inSPIN, located at the Department of Molecular Biology (Yonathan Sonntag and Poul Nissen), the Department of Chemistry (Maria Musgaard and Birgit Schiøtt), the Department of Physiology and Biophysics (Claus Olesen and Jesper Vuust Møller) and the Bioinformatics Centre – all at Aarhus University in Denmark (Lea Thøgersen).
Link to the article:
Mutual adaptation of a membrane protein and its lipid bilayer during conformational changes
Yonathan Sonntag1,6, Maria Musgaard2,5, Claus Olesen3,5, Birgit Schiøtt2,6, Jesper Vuust Møller3,5, Poul Nissen1,5, Lea Thøgersen1,4,5
1Department of Molecular Biology, Aarhus University, Denmark
2Department of Chemistry, Aarhus University, Denmark
3Department of Physiology and Biophysics, Aarhus University, Denmark
4Bioinformatics Research Centre, Aarhus University, Denmark
5Centre for Membrane Pumps in Cells and Disease – PUMPKIN, Danish National Research Foundation, Aarhus University, Denmark
6Centre for Insoluble Protein Structures – inSPIN, Danish National Research Foundation, Aarhus University, Denmark
More information
Associate Professor Lea Thøgersen
Bioinformatics Research Centre and Department of Molecular Biology, Aarhus University, Denmark
lea@birc.au.dk, mobile: +45 3026 1236
PhD student Maria Musgaard
Department of Chemistry, Aarhus University, Denmark
musgaard@chem.au.dk, mobile: +45 5190 5887
Professor Poul Nissen
Department of Molecular Biology, Aarhus University, Denmark
pn@mb.au.dk, mobile: +45 2899 2295
www.pumpkin.au.dk
Text: Lea Thøgersen, Poul Nissen and Lisbeth Heilesen
Translation: Lisbeth Heilesen
