An enzyme complex maintains genome stability at replication roadblocks
During each cell cycle, the genome must undergo complete and faithful DNA replication to allow correct segregation of the duplicated chromosomes to the daughter cells. However, imperfections in the DNA template or physical roadblocks can threaten the fidelity of the replication machinery and thereby jeopardise genomic integrity. Studies on how cells cope with replication dangers are thus crucial for understanding how cells fight genomic instability.

To analyse the cellular response to DNA roadblocks without treating cells with non-specific genotoxic agents leading to high-levels of non-physiological replication fork stalling events, the researchers took advantage of the ribosomal DNA (rDNA), which contains natural replication barriers called RFBs (Replication Fork Barrier). These RFBs become active upon binding of a protein called Fob1.
The researchers engineered a cellular system in baker’s yeast (S. cerevisiae), where the Fob1 expression was controlled by an inducible promoter. The system (Fob-block) allows them to strongly induce replication fork barriers in the form of a non-covalent protein-DNA complex either at their natural location within the rDNA, or when placed in a chromosomal context outside the rDNA.
Using this system, the researchers uncovered a pivotal non-enzymatic role for a highly conserved complex, the MRX complex (Mre11, Rad50, Xrs2) at the replication fork barriers. The MRX complex is known to be involved in the repair of DNA double strand breaks and is also engaged in the cell’s surveillance mechanism called checkpoint. Inherited mutations in the human counterpart, the MRN complex (Mre11, Rad50 and Nbs1), is associated with human genome instability syndromes like ataxia-telangiectasia-like disease (ATLD), NBS and NBS-like disorder (NBSLD). For example, NBS patients have an extremely high rate of malignancies (approx. 40% before the age of 30), which really highlights the importance of the Mre11 complex for maintaining genome integrity and provides impetus to the researchers attempts at understanding the molecular action of this complex.
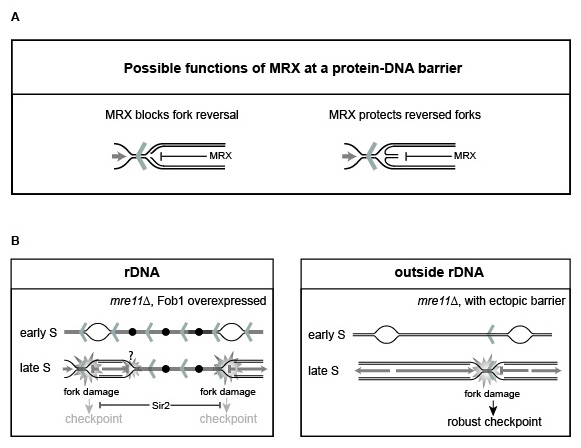
The Fob-block system unravels a new role of the MRX complex for genome maintenance when replication forks stall due to impediments, which relies on the structural integrity of the complex and not its enzymatic activity. The researchers show that the MRX complex protects stalled replication forks, as absence of a structural competent MRX complex leads to abnormal processing of the DNA giving rise to single-stranded DNA.
Interestingly, their studies unravel a hitherto unknown phenomenon showing that abnormal DNA structures generated at replication fork barriers in the absence of a structural competent MRX complex do not activate the cellular checkpoint when this occurs within the rDNA, whereas abnormal structures generated at a protein-DNA barrier outside the rDNA gives a strong checkpoint response. They were able to show that the absence of a checkpoint within the rDNA is due to altered chromatin structure in this cellular subcompartment. This result suggests that the checkpoint is governed by chromosomal context, which may also play significant roles in other chromosomal contexts.
Link to the article in Nucleic Acids Research:
Further information
Associate Professor Lotte Bjergbæk
Department of Molecular Biology and Genetics
Aarhus University, Denmark
lbj@mb.au.dk – +45 2326 2695
4.2.2013
