How human gene promoters become directional
An international research team has disclosed how cellular gene transcription and RNA degradation processes collaborate to achieve a directional output from human genes.
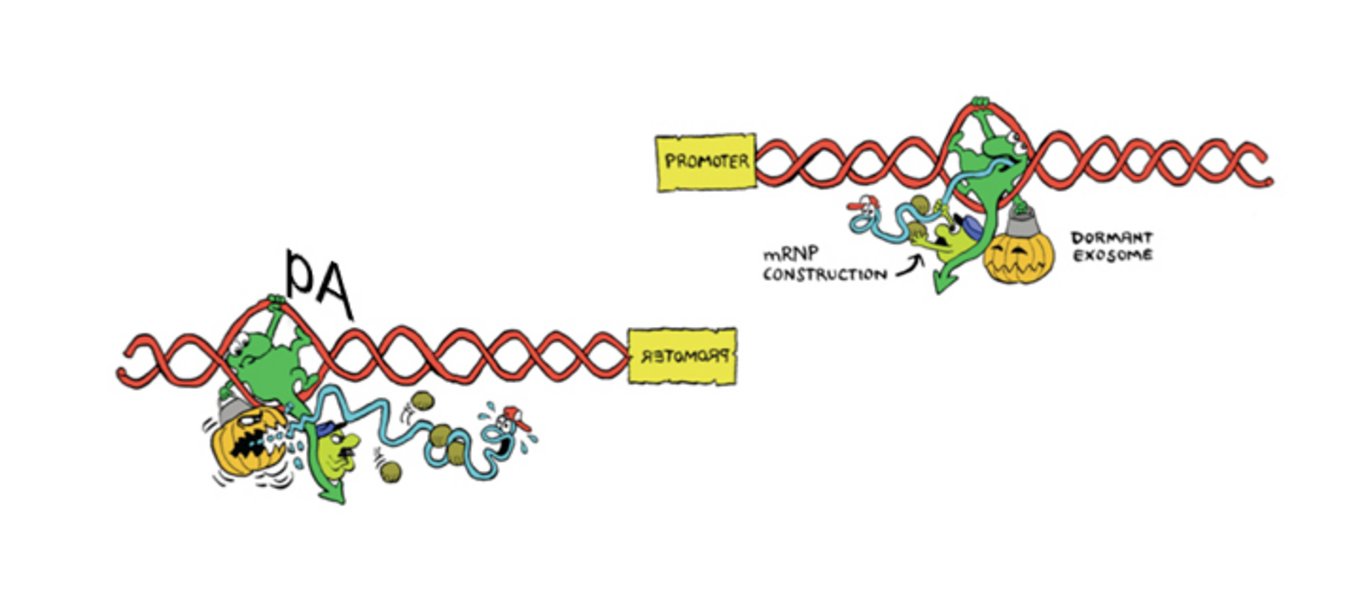
Human gene promoters are inherently bidirectional yet their RNA output is strictly unidirectional – how can that be? Back in 2008, researchers at Aarhus University uncovered hidden layers of promoter upstream transcription (producing so-called PROMoter uPstream Transcripts (PROMPTs)) by depleting cells of the major nuclear exoribonucleolytic protein complex, the RNA exosome (Preker et al., Science 2008).
However, the question of how PROMPTs are rapidly cleared, while their neighbouring mRNAs are retained as functional (protein-coding) entities, has remained. New research published online this week at Nature Structural and Molecular Biology now provides the answer.
PROMPTs are short and capped non-coding RNAs produced upstream of all active human gene promoters. To unravel the mechanism of their transcription termination and linkage to exosome-mediated degradation, Postdoctoral Researcher Evgenia Ntini from Torben Heick Jensen’s group at Aarhus University engaged in a collaboration with Lars Steinmetz’s group at the EMBL in Heidelberg as well as Albin Sandelin’s group at the BRIC in Copenhagen.
Together the researcher used a RNA high-throughput sequencing approach combined with in depth bioinformatics analysis to acquire an unprecedented deep and genome-wide picture of PROMPTs, allowing their comprehensive sequence motif analysis. This effort revealed that PROMPTs are produced antisense to RNA polymerase II transcribed genes, and they arise due to a strong bi-directional gene promoter activity.
How then is PROMPT transcription termination ensured and why are these transcripts targeted for rapid exosome-mediated degradation? Sequence motif searches around PROMPT 3’ends demonstrated a motif composition, which is surprisingly similar to that of mRNA 3’ends, including the presence and well-defined positional peaks of the hexameric AWTAAA sequence. Thus, PROMPT transcripts appear to terminate in a way, which is remarkably similar to that of mRNAs and by utilisation of polyadenylation-like signals. Importantly, however, upstream-antisense PROMPT 3’ends – and therefore their poly(A)-like sites – are concentrated within a fairly narrow distance window from their start sites.
This configuration makes the resulting transcripts highly unstable, and as proximal poly(A) sites are selected against in the ’mRNA direction’ of transcription, such asymmetric sequence distribution around gene promoters explains why the RNA output from the transcription process becomes highly directional. In other words, cellular gene transcription and RNA degradation processes collaborate to achieve a directional output from human genes.
Further investigation
The investigators speculate that the promoter-proximity per se of PROMPT poly(A)-sites render them exosome sensitive. On-going efforts are now directed towards understanding whether this disclosed mechanism is also operational elsewhere in the human genome. This is an extremely important question as > 90% of the human genome is transcribed and precautions need to be in place to suppress the non-functional portion of such a massive RNA output.
The work was conducted by Postdoctoral Researcher Evgenia Ntini and other members of Torben Heick Jensen’s group at the Danish National Research Foundation-funded Centre for mRNP Biogenesis and Metabolism, Department Molecular Biology and Genetics, Aarhus University, in collaboration with the groups of Lars Steinmetz at the EMBL in Heidelberg and Albin Sandelin at the BRIC in Copenhagen.
Link to the research article in Nature Structural & Molecular Biology: Polyadenylation site–induced decay of upstream transcripts enforces promoter directionality.
Further information
Professor Torben Heick Jensen
Department of Molecular Biology and Genetics
Centre for mRNP Biogenesis and Metabolism
Aarhus University, Denmark
thj@mb.au.dk - +45 6020 2705
