Millions of small RNAs paint a bigger picture of human gene activity
Scientists have discovered the origin of a new group of small RNAs (sRNAs) that contain genetic information. This discovery can lead to a better understanding of how human genes are controlled, and with this knowledge, scientists might one day be able to regulate some of the genes that cause various diseases.

The international research team has provided plausible explanations for the origin of an abundant class of sRNAs, molecules that have puzzled the scientific community since their original discovery. In their study, published in this week’s edition of the renowned journal Nature Structural and Molecular Biology, the researchers show that sRNAs originating from human genes are largely generated as degradation intermediates of longer RNAs. They also provide evidence supporting which enzymes are responsible for this degradation, and these may prove to be new key players in the regulation of human gene activity in the future.
Background information
It used to be so easy. For decades, ribonucleic acid (RNA) was regarded as a polymeric molecule, one of whose roles was to serve as a messenger (hence commonly called mRNA) that transmits the genetic information contained in DNA inside the cell nucleus to the outside as a translation template for the production of proteins. For its own production, in a process known as transcription, mRNAs rely on a molecular machine, the so-called RNA polymerase II (RNAPII).
Recent technological advances now enable investigations in unprecedented depth into the total cellular RNA content. Two of the most burning questions have been the origin and purpose – if any – of a highly abundant and diverse set of RNAs that, like mRNAs, are derived from genes, but unlike mRNAs are much shorter. These sRNAs are only up to around 25 nucleotides long, while mRNAs can easily reach lengths in the thousands.
Small RNAs: nothing but driftwood?
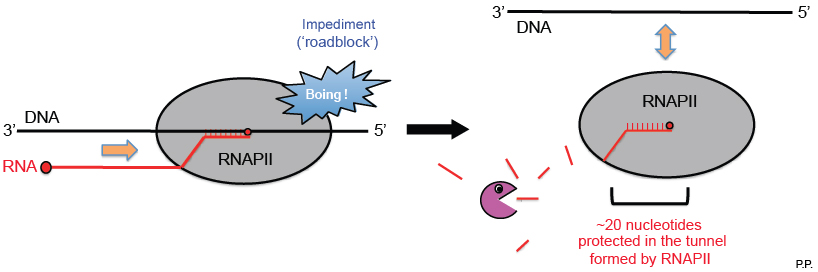
By investigating millions of sRNAs, some from cells depleted of RNA degradation enzymes (e.g. Xrn1 and Xrn2) to make degradation intermediates more stable, and others from specific cellular compartments, the likely origins of the sRNAs were pinpointed. To gain more mechanistic insight, these findings were then compared with previously published maps of the position of RNAPII and the large DNA structures, nucleosomes. As mRNA is produced, its front end passes through a tunnel formed by RNAPII itself. Interestingly, the peak length of the sRNAs (around 20 nucleotides) corresponds exactly to the number of nucleotides that the tunnel can accommodate. These results paint a picture where a sizable fraction of sRNAs are generated when transcription is aborted and the unfinished mRNA is attacked by degrading enzymes while still attached to RNAPII, leaving behind only those approximately 20-nucleotide fragments that are sheltered in the tunnel (see Figure).
It has been known for a while that transcription is especially prone to early failure during the event, and indeed it is here where many sRNAs originate. The scientists could also identify many sRNAs that stem from sites further down the gene as by-products of a process known as splicing, another event that is critical for the generation of most functional mRNAs, or as remnants of defects in this process.
As icing on the cake, the team identified not only some of the sRNAs associated with splicing, but also sRNAs originating from the very ends of protein-coding genes as rare subclasses of sRNAs that show promises of being functional as they were associated with the Argonaute proteins that mediate the gene-regulatory functions of microRNAs. As a whole, these new observations support the notion that many mRNAs remain unfinished and are rapidly discarded instead. Rather than being merely wasteful, however, it is thought that this process is one of the many ways by which cells shield themselves against the potentially deleterious effects of faulty mRNAs, true to the precautionary principle “better safe than sorry”.
The scientists now look forward to studying these mechanisms in greater detail by both further genome-wide studies and more rigorous investigation of selected genes – efforts they hope will reveal how RNAPII is controlled during early transcription.
The research was carried out equally by the groups headed by Professor Torben Heick Jensen, Director of the Centre for mRNP Biogenesis and Metabolism, Department of Molecular Biology and Genetics, Aarhus University, and Associate Professor Albin Sandelin, Bioinformatics Centre, Department of Biology, and the Biotech Research and Innovation Centre (BRIC), University of Copenhagen. The first authorship was split three ways between Postdoctoral Fellow Eivind Valen (BRIC), Postdoctoral Fellow Pascal Preker and Graduate Student Peter R. Refsing (both from the Centre for mRNP Biogenesis and Metabolism). Professor Gunter Meister and members of his group at the Department of Biochemistry, University of Regensburg, joined them in their effort.
Further information
Professor Torben Heick Jensen
Centre for mRNP Biogenesis and Metabolism
Department of Molecular Biology and Genetics, Aarhus University, Denmark
thj@mb.au.dk, +45 6020 2705
Text: Peter Refsing, Torben Heick Jensen and Lisbeth Heilesen
Translation: Lisbeth Heilesen
