Prey digestion by the carnivorous Venus flytrap
A newly published study by researchers from Aarhus University provides the most comprehensive analysis to date of the protein composition in the digestive juice of a carnivorous plant, and this contributes significantly to the understanding of prey digestion in these plants. The identified, unique digestive enzymes identified by the researchers could be a source of inspiration for the enzyme industry.

The carnivorous plant Venus flytrap
The Venus flytrap (Dionaea muscipula) (Figure 1) is one of the most well-known carnivorous plants because of its unique ability to capture small animals, usually insects or spiders, through a special snap-trapping mechanism. The plant actively traps the prey, which is killed and digested to assimilate nutrients as the plants normally grow in mineral-deficient soils.
The plant has fascinated both scientists and laymen for years, and in his book ”Insectivorous Plants” published in 1875 Charles Darwin described the plant as ”one of the most wonderful in the world”.
The trapping mechanism has been studied in detail, but prior to the published study, knowledge about the digestion mechanism was limited, not only for the Venus flytrap, but for carnivorous plants in general. In addition, the study contributes to a better understanding of the evolution of carnivorous plants and has the potential of revealing enzymes of industrial relevance.
Identification of proteins in digestive juice
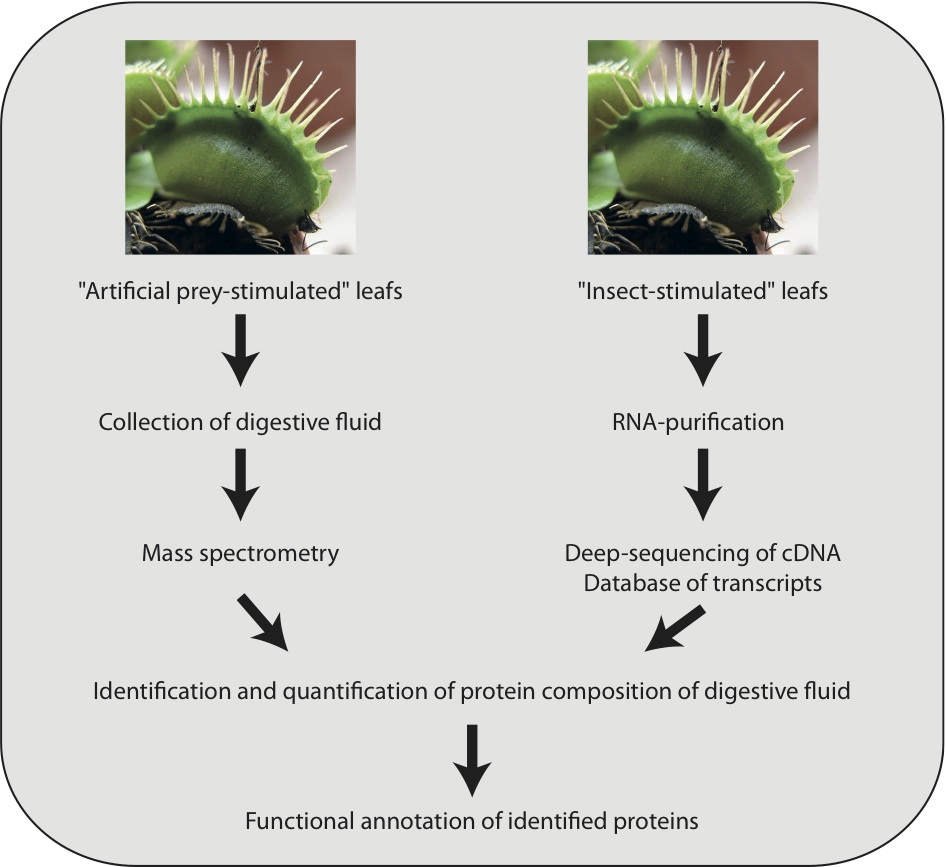
To identify and quantify the proteins present in the digestive fluid, an approach combining transcriptomics and proteomics was applied (Figure 2). The researchers initially carried out deep sequencing of cDNA derived from Venus flytrap leaves that were stimulated by different methods, including feeding with ants and beetles. For the proteomics analyses, protocols were developed that avoided contamination of digestive fluid with prey proteins. Using these artificial stimulation methods, the researchers collected digestive fluid and analysed it by mass spectrometry.
The data obtained were searched against the transcriptomes and the proteins present in the digestive fluid were identified, quantified, and functionally annotated using this approach. The data showed that the protein composition is very different from that in animal digestive juices. In vertebrates and in the carnivorous plant Nepenthes, the proteolytic enzymes are mostly aspartic proteases. In the Venus flytrap, however, cysteine proteases were the most abundant class, followed by a serine carboxypeptidase and aspartic proteases. As expected, chitinases that probably facilitate degradation of the exoskeleton of trapped insects were also identified. The majority of the most abundant proteins in the digestive fluid were homologous with proteins involved in the defense against pathogens in other plants. This suggests a shift from defense-related processes to digestion-related processes among the carnivorous plants.
The majority of the most abundant proteins in the digestive fluid were homologous with proteins involved in the defense against pathogens in other plants. It suggests a shift from defense-related processes to digestion-related processes among the carnivorous plants.
The strategic research alliance NOVENIA
Dr. Jan J. Enghild’s group is continuing its work on the Venus flytrap plant and recombinant versions of the major enzymes to gain a better understanding of their biochemical characteristics and the digestion mechanism at the molecular level. In addition, the biochemical analyses may reveal properties that could be exploited commercially. This is of interest for the industrial partners in the research alliance ‘Novel enzymes of industrial relevance: specialized proteolytic enzymes for release of new bioactive peptides (NOVENIA)’, see www.novenia.dk, supported by the Danish Council for Strategic Research. In NOVENIA, different digestive systems and marine microbial communities are being analysed to identify proteolytic enzymes of industrial relevance.
The findings have just been published in Molecular and Cellular Proteomics (http://www.mcponline.org).
The project was carried out in collaboration with colleagues at the University of Southern Denmark, the Max Planck Institute of Molecular Plant Physiology (Potsdam), the Heinrich Heine University Düsseldorf, and the University of Würzburg.
Waltraud X. Schulze1, Kristian W. Sanggaard2, Ines Kreuzer3, Anders D. Knudsen2, Felix Bemm3, Ida B. Thogersen2, Andrea Brautigam4, Line R. Thomsen2, Simon Schliesky4, Thomas F. Dyrlund2, Maria Escalante-Perez3, Dirk Becker3, Jorg Schultz3, Henrik Karring5, Andreas Weber4, Peter Hojrup5, Rainer Hedrich3 and Jan J. Enghild2*
1Max Planck Institute of Molecular Plant Physiology, Germany;
2Aarhus University, Denmark;
3 University of Wuerzburg, Germany;
4 Heinrich Heine University, Germany;
5 University of Southern Denmark, Denmark
*Corresponding author; email: jje@mb.au.dk
More information
Kristian Wejse Sanggaard
Department of Molecular Biology and Genetics
Aarhus University
Denmark
krs@mb.au.dk; Cell phone: +45 2374 1497
Text: Kristian Wejse Sanggard and Lisbeth Heilesen
