Discovery of connection between RNA splicing and decay machineries
RNA synthesis, splicing and degradation are key activities in eukaryotic gene expression regulation. A collaborative effort between researchers from the Max Planck Institute, Martinsried and Aarhus University now reveals the physical basis for linking RNA degradation to the splicing process.
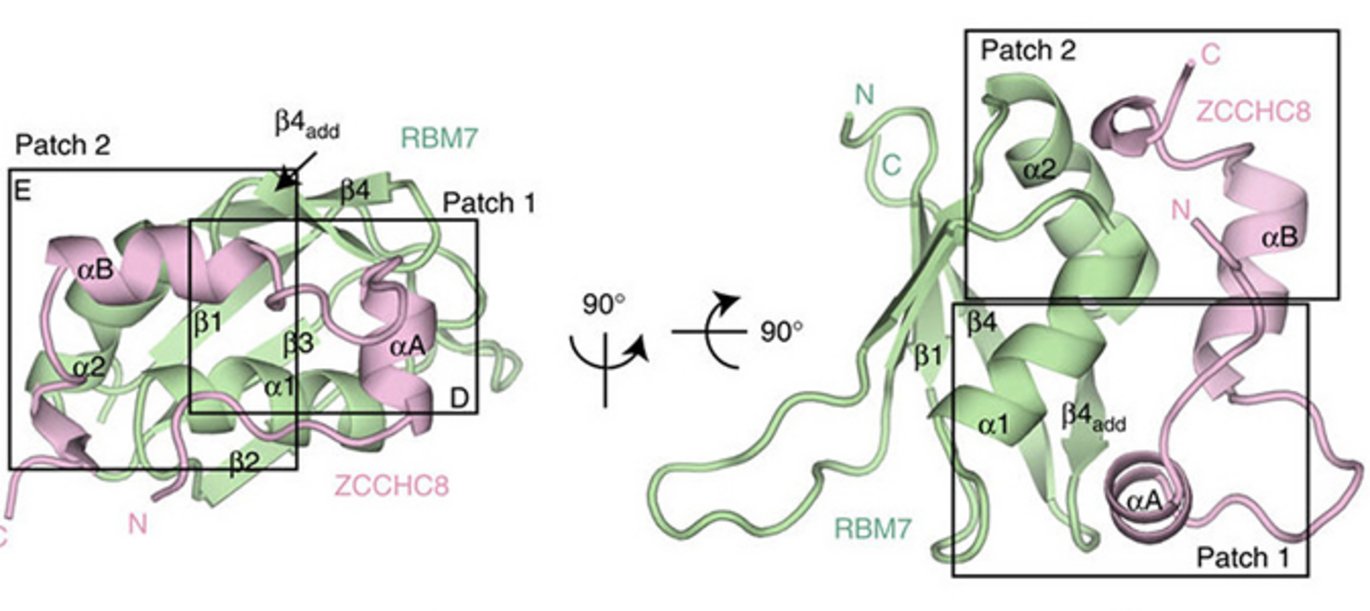
Many eukaryotic RNA species undergo nuclear splicing, both to produce mature messenger RNA (mRNA) and to liberate introns. Since introns often harbor smaller RNA species that need to be ‘processed out’, degradation/processing factors are required – these enzymes also serve to conduct complete intron degradation. In the nucleus, the major RNA degradation machinery is the so-called RNA exosome. In vivo, exosome activity is regulated by associated enzymatic co-factors and RNA-binding adapters, which facilitate exosome access and recruitment to a multitude of substrates.
A few years ago, the laboratory of Torben Heick Jensen at the Department of Molecular Biology and Genetics, Aarhus University, identified and characterized an essential exosome co-factor, the ‘Nuclear EXosome Targeting (NEXT) complex’ which recruits short RNA targets to the active sites of the RNA exosome. NEXT is a trimeric complex composed of the RNA recognition motif (RRM)-containing RBM7 and the zinc-knuckle ZCCHC8 proteins forming a dimer associating with the hMTR4 RNA helicase that is also engaged with other exosome cofactors*Interestingly, proteomic and transcriptomic studies using RBM7 as a bait suggested a possible connection between RBM7 and the spliceosomal SF3b complex**
In collaboration with Prof. Elena Conti’s laboratory at the Max-Planck Institute of Biochemistry in Martinsried, Germany, the interacting regions between RBM7 and ZCCHC8 were narrowed down to the RRM domain of RBM7 (RBM7-RRM) and a proline-rich region of ZCCHC8 (ZCCHC8-Pro). The crystal structure of the RBM7-ZCCHC8 core complex of NEXT was successfully resolved (see Figure) and the amino acid residues engaged in the interaction were determined and validated, by residues mutations, in vitro and in vivo.
Surprisingly, bioinformatics analysis showed remarkable sequence similarities between RBM7-RRM and ZCCHC8-Pro with two proteins of the spliceosomal SF3b complex, SAP49 and SAP145, respectively. Indeed, interaction studies revealed direct physical contact between RBM7-RRM and SAP145-Pro, thus linking RBM7 to RNA splicing factors. Although RBM7-RRM binding to either the proline-rich domains of ZCCHC8 or SAP145 turned out to be mutually exclusive, the potential ability of RBM7 to homo-dimerize might reflect a possible concomitant link between exosome-mediated degradation and splicing. One functional outcome of this connection appears to be the targeting of the exosome to introns in order to excise intron-hosted snoRNAs. Other functional consequences of NEXT-SF3b interaction are presently being investigated.
The research project was carried out by postdoc Sebastian Falk and PhD student Ksenia Finogenova from the laboratory of Elena Conti at the Max-Planck Institute of Biochemistry, Martinsried, Germany in collaboration with postdoc Mireille Melko from the laboratory of Torben Heick Jensen, Aarhus University, Denmark. The paper is published in Nature Communications.
*Lubas et al. Mol Cell 2011, Meola et al. Mol Cell 2016.
**Lubas et al. Mol Cell 2011, Lubas et al. Cell Reports 2015
For further information, please contact
Postdoc Mireille Melko og Professor Torben Heick Jensen
Institut for Molekylærbiologi og Genetik, Aarhus Universitet
melko@mbg.au.dk; thj@mb.au.dk - mobil: 6020 2705
