Finding hidden treasures in our DNA
The human genome is promiscuously transcribed yielding RNA from >75% of its DNA, and throughout the years, researchers world-wide have tried to find out how much of this material is functional. Danish researchers have now received a prestigious grant from the Lundbeck Foundation to address this problem.
In addition to its ~20.000 protein-coding genes, the human genome harbours an even larger number of genes encoding so-called ‘non-coding RNAs’ (ncRNAs). While some ncRNAs are clearly critical to cells, there is an ongoing and heated debate about how many have clear molecular functions. This is an important question, because if just a fraction of these molecules is functional, the number of cellular genes would rise dramatically.
As part of cellular quality control, spurious transcripts are constantly removed, and only a subset of ncRNAs will retain copy numbers compatible with function. Finding these, however, is a major challenge and efficient strategies are warranted. Moreover, the ability of pervasive transcription to generally regulate gene expression is tremendous but not adequately investigated.
Professor Torben Heick Jensen has now received a prestigious grant from the Lundbeck Foundation amounting to DKK 10 million over five years to conduct the research project “Discovery of new genes and regulatory paradigms”. The project, part of which is a collaborative effort with Professor Albin Sandelin’s group from the Department of Biology, Copenhagen University, aims to reveal new functional entities and regulatory principles within mammalian genomes.
In the present project relevant model systems will be coupled to high throughput biochemical and computational methods to further the identification of new ncRNA. A series of screening steps will subsequently pinpoint and categorize examples for further mechanistic studies. Importantly, this will also expose when activities, other than the ncRNA itself (for example regulatory transcription events), are functionally relevant.
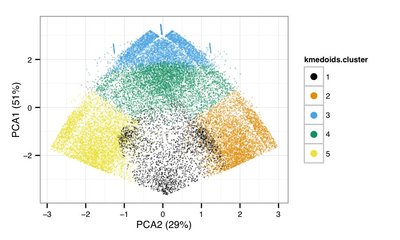
Researchers at the University of Copenhagen and Aarhus University have previously devised a combined experimental and computational method, which classifies ncRNAs based on their biochemical properties inside human cells (see Figure). The strategy relies on a systematic categorisation of human RNAs by the nature of their transcription initiation, by their overall expression levels and by their sensitivity to the RNA-degrading exosome complex. These measures are surprisingly effective at correctly classifying annotated transcripts, including ncRNAs of known function.
Although the project revealed that most ncRNAs are more labile than protein-coding mRNAs, it also identified uncharacterised stable ncRNAs, hidden among a vast majority of unstable transcripts. This implies that ncRNAs with possible functions can be pinpointed for further analyses. Within the scope of the Lundbeck grant, this strategy will be expanded to other model systems, like mouse embryonic stem cells.
For further information, please contact
Professor Torben Heick Jensen
Department of Molecular Biology and Genetics
Aarhus University, Denmark
+45 6020 2705, thj@mbg.au.dk
