New insights into fundamental stress-regulated cellular processes
Danish and Canadian researchers have uncovered important molecular details about the regulation of the cell biosynthesis-machinery during cellular stress. This knowledge has implications for anti-cancer treatment, since the implicated factors are key regulators of cell growth and proliferation.
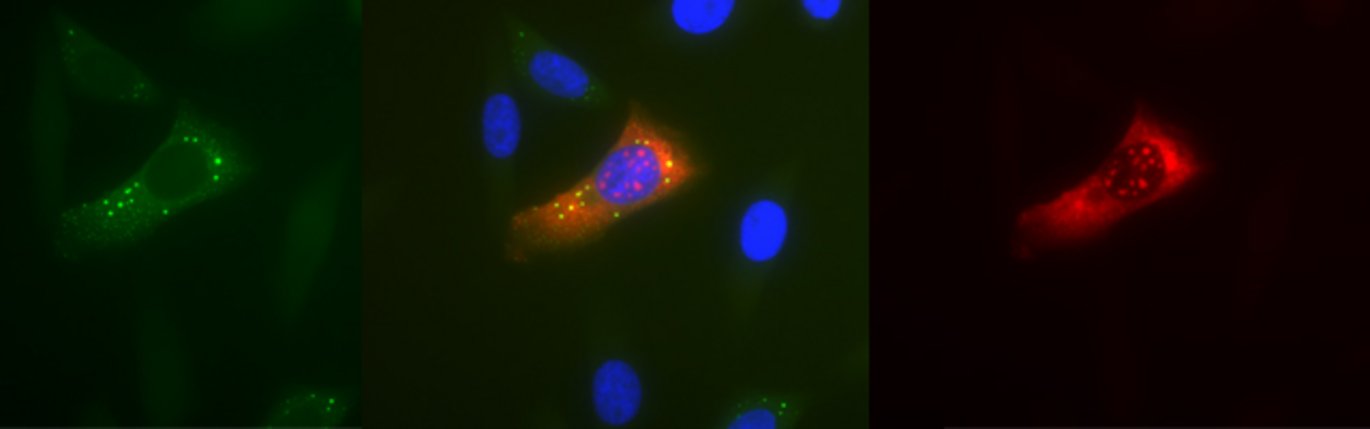
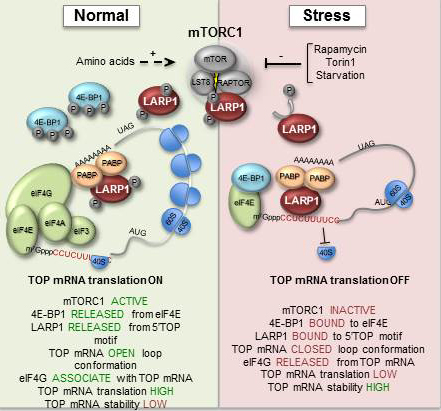
When cells become exposed to stress, such as amino acid starvation, hypoxia or heat-shock, gene-regulatory events are immediately reprogrammed by a process that largely controls the cell biosynthesis-machinery including ribosomes. Ribosomes are macromolecular ’machines’ that translate the genetic code, embedded in messenger RNA (mRNA), into protein. During stress, a large class of mRNAs, harbouring a unique sequence element (TOP mRNA), is selectively excluded from the ribosomes (see figure 2), which allows the cell to survive and cope with the harsh conditions. This delicate process is controlled by a number of cellular signals that converge at the central mTOR (mammalian Target Of Rapamycin) signalling pathway.
The study has shown how the mTOR signalling pathway relays its signals all the way to the ribosome, by identifying a hitherto unknown substrate for mTOR, LARP1, which in turn specifically inhibits TOP mRNA translation during stress (see figure 2).
mTOR and LARP1 downregulate translation to lower energy consumption during stress
During normal conditions, LARP1 is phosphorylated by the active mTOR kinase, which in turn regulates the ability of LARp1 to bind TOP mRNA (left panel of figure). When mTOR is deactivated by cellular stress, LARP1 is no longer highly phosphorylated, which increases its TOP mRNA binding affinity and translational repression (right panel of figure 2).
The study has also provided novel mechanistic insights into this process. It turns out that LARP1 competes with an essential translation initiation factor, eIF4G, for association with TOP mRNA. When mTOR is inactivated, LARP1 ’wins’ this battle, which leads to co-repression of hundreds of TOP mRNAs. This allows the cell to mount a proper stress-response and ensure its survival.
A large number of mTOR drugs are currently being tested in various anti-cancer clinical trials, since mTOR stimulates cell growth and proliferation. Given the druggable nature of the mTOR signaling pathway and the prominent roles for ribosome biogenesis, protein synthesis and mRNA decay in cancer, we anticipate that the LARP1 status of cancer cells may be a valuable marker for determining anti-cancer drug (i.e. rapamycin) resistance.
The scientific article was recently published in the Journal of Biological Chemistry:http://www.jbc.org/content/early/2015/05/04/jbc.M114.621730.long
More information
Associate Professor Christian Kroun Damgaard
Department of Molecular Biology and Genetics
Aarhus University, Denmark
ckd@mbg.au.dk - +45 2970 0599
