Newly discovered RNA decay pathway inside human nuclei
Genomes are promiscuously transcribed into RNA. However, not all of this material is immediately useful, which means it has to be targeted and degraded in order to sustain cellular life. A newly discovered RNA decay pathway functioning inside human nuclei does just that.
Our genomes are hyperactive, producing a variety of transcripts. A chief component of this so-called ‘pervasive transcription’ is non-coding RNA species, many of which are degraded soon after their production. So why are such ‘ghostly’ RNAs made at all? Given their usually short half-lives, they are unlikely to all be functional. However, this genome activity may be important for other aspects of gene expression or play a role as a reservoir for evolution to create new functional transcription units. Regardless, these RNAs need to be turned over rapidly not to cause a catastrophic built up, disrupting or delaying other vital cellular decision.
RNAs destined for degradation are diverse – short and long, immature and mature - so how are they identified? Unstable RNA species are often degraded in eukaryotic nuclei by the ribonucleolytic RNA exosome, which is well conserved from yeast to human. In doing so, the RNA exosome utilises adaptor complexes, guaranteeing target specificity and the consequent removal of the intended molecules.
A few years ago the laboratory of Torben Heick Jensen at the Department of Molecular Biology and Genetics, Aarhus University, discovered and characterised a nuclear exosome adaptor, called ‘the Nuclear EXosome Targeting (NEXT)’ complex, which recruits the RNA exosome to short and primarily unprocessed RNA species*. This finding highlighted the possibility that another adaptors might exist to target longer and more processed RNAs. A simple, yet effective, idea, which led to the present discovery of the so-coined ‘PolyA eXosome Targeting (PAXT)’ connection.
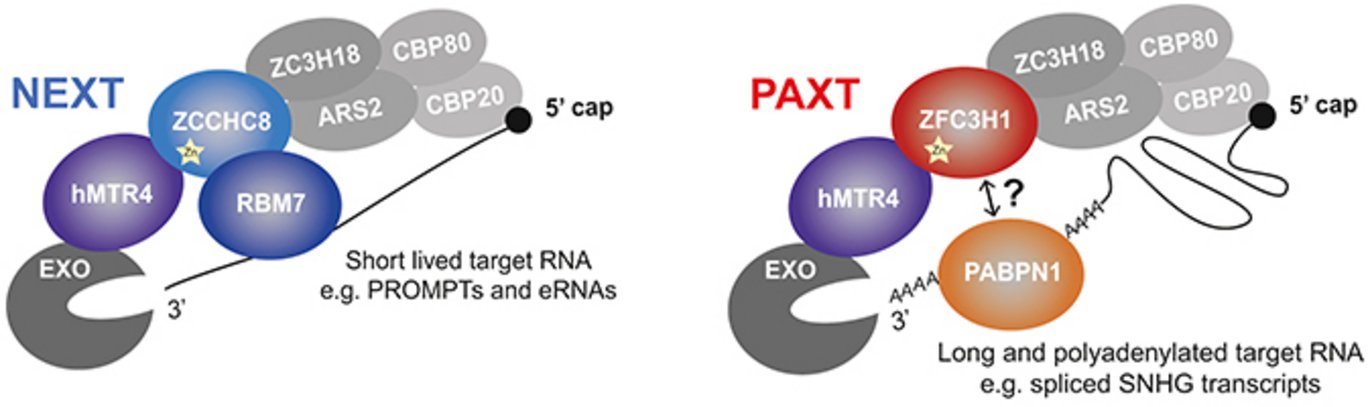
PAXT protein composition mimics to some extent that of NEXT by also harbouring the RNA helicase hMTR4, which is the fundamental and direct physical link to the RNA exosome (see Figure). Moreover, PAXT harbours a zinc finger protein, ZFC3H1 (while NEXT harbours the zinc finger protein ZCCHC8), and the polyA-tail binding protein PABPN1 (NEXT harbours the RNA-binding protein RBM7) that guarantees PAXT’s recognition of mainly polyadenylated transcripts.
Despite these similarities, PAXT and NEXT interact with the RNA exosome in a mutually exclusive manner. That PAXT and NEXT pathways are functionally distinct was also demonstrated by high throughput RNA-sequencing analysis. In particular, PAXT was revealed to target polyadenylated transcripts, which are also significantly longer than classical NEXT substrates. In addition to such ability to ‘monitor’ the processing status of nuclear RNA, both PAXT and NEXT pathways appear to be capable of detecting capped RNA by virtue of their physical linkages to the cap-binding complex (CBC) (see Figure). The research group is now trying to reveal other biochemical aspects discriminating PAXT and NEXT substrates as well as hunting for additional and yet-to-be-discovered RNA exosome adapters.
The published results, spearheaded by postdocs Nicola Meola and Michal Domanski as well as PhD student Evdoxia Karadoulama, have been published in Molecular Cell. They were achieved via collaboration with researchers from the Universities of Copenhagen and Southern Denmark.
*Lubas et al. Mol. Cell 2011; Andersen et al. Na.t Struct. Mol. Biol. 2013; Lubas et al. Cell Reports 2015.
For further information, please contact
Professor Torben Heick Jensen
Department of Molecular Biology and Genetics
Centre for mRNP Biogenesis and Metabolism
Aarhus University, Denmark
thj@mb.au.dk - mobile: +45 60202705
