Researchers reveal dual role for human RNA decay factor
Researchers at Aarhus University have characterized the human RNA decay factor ZC3H18 and discovered its unanticipated role in the production of protein-coding RNA. The new study, published this week in <em>Cell Reports</em>, therefore reveals a double-faced activity of ZC3H18 in nuclear RNA fate decisions.
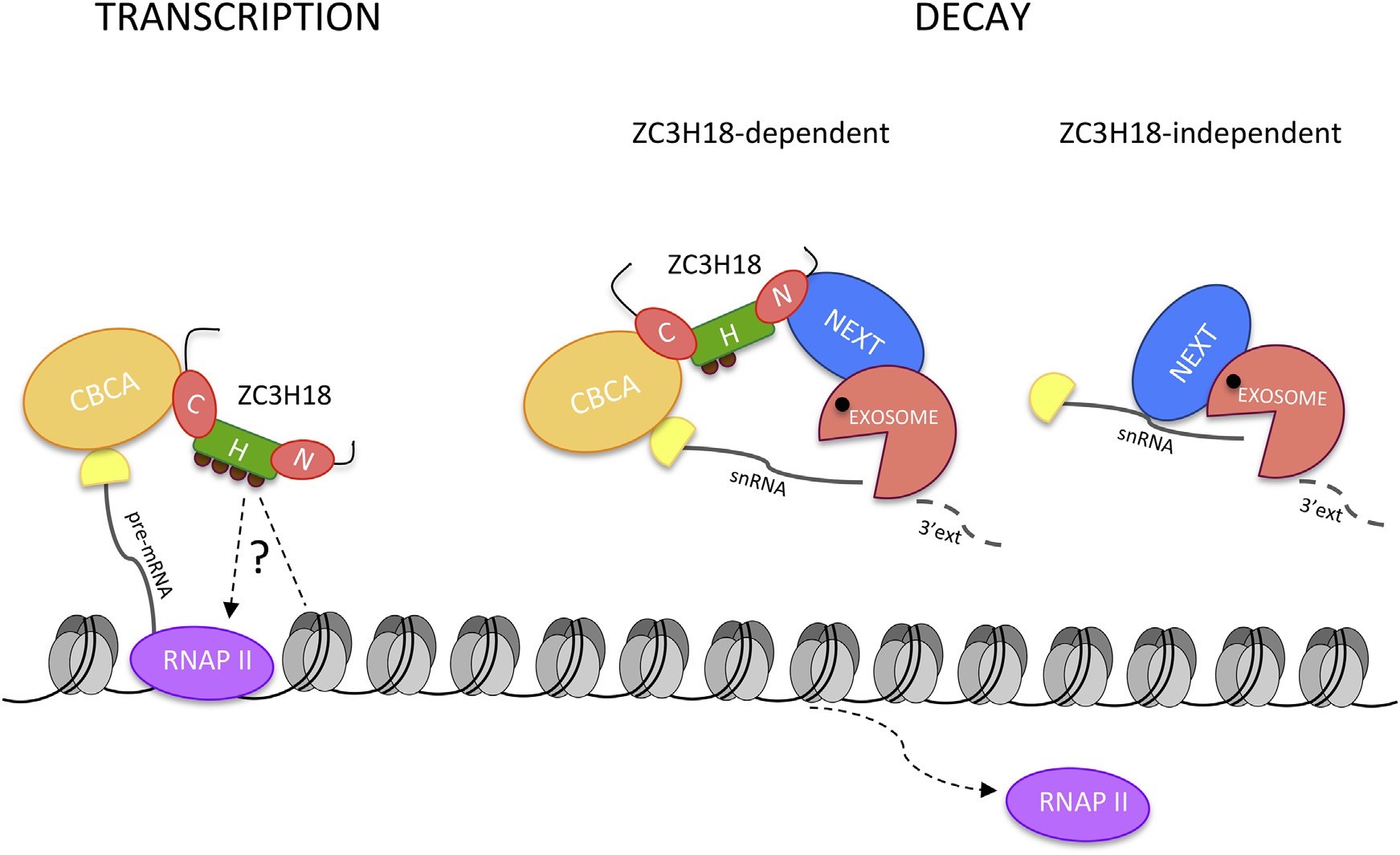
Eukaryotic genomes are pervasively transcribed leading to a multitude of RNAs, such as protein-coding RNAs, non-coding RNAs and various cryptic RNAs, which are most often rapidly degraded. Additionally, newly synthetized RNAs undergo maturation to become functional molecules – a process generating even more spurious RNA by-products. Unless kept under tight surveillance, such massive RNA production will jeopardize cell survival, and it is therefore fortunate that mechanisms exist ensuring that proper levels of functional transcripts are maintained. This is governed by specialized protein complexes, which control RNA production and destruction pathways. The new study spearheaded by postdoc Kinga Winczura underscores that the zinc-finger protein ZC3H18 has a foot in each camp affecting both RNA productive and destructive processes.
In order to understand how RNA metabolism is regulated, it is necessary to investigate how proteins involved in the process interact with each other. The uncharacterized zinc finger protein ZC3H18 binds to the cap-binding complex (CBC), present at the beginning (cap) of many types of RNAs, while also associating with the so-called nuclear exosome targeting (NEXT) complex, linking to RNA decay (Figure, right panel).
Kinga Winczura from Torben Heick Jensen’s laboratory at the Department of Molecular Biology and Genetics at AU collaborated with researchers at the University of Southern Denmark in Odense and the Rockefeller University in New York to perform an extensive biochemical and functional characterization of ZC3H18. Surprisingly, a new phosphorylation-dependent isoform of ZC3H18 was discovered, which associates with histone proteins that wrap around the DNA of our cells and are crucial factors during RNA synthesis as they bear special marks, which can promote or inhibit gene transcription (Figure, left panel).
Additional examination of ZC3H18 function, using genome-wide analyses, demonstrated its impact on transcription of a subset of protein-coding genes. This activity was shown to require the CBC-interacting domain of the protein, with the activity of some genes being also dependent on the NEXT- and/or histone-interacting domains. Exactly how ZC3H18 selects RNAs for these antagonistic destinies is presently unclear and remains a fascinating focus for future investigation.
For further information, please contact
Professor Torben Heick Jensen
Department of Molecular Biology and Genetics
Aarhus University, Denmark
thj@mbg.au.dk - +45 60202705
