Sorting RNA for production or decay
Our genomes are promiscuously transcribed into RNA. How cells manage to sort this massive genomic output into functional and non-functional material has remained enigmatic. New research describes protein interactions involved in such RNA fate determination.
Genomes are hyperactive, producing RNA from 80-90% of their DNA. Since not all of this material has cellular half-lives compatible with function, it follows that a complicated RNA sorting process must take place, leaving only molecules sustaining cellular maintenance and growth.
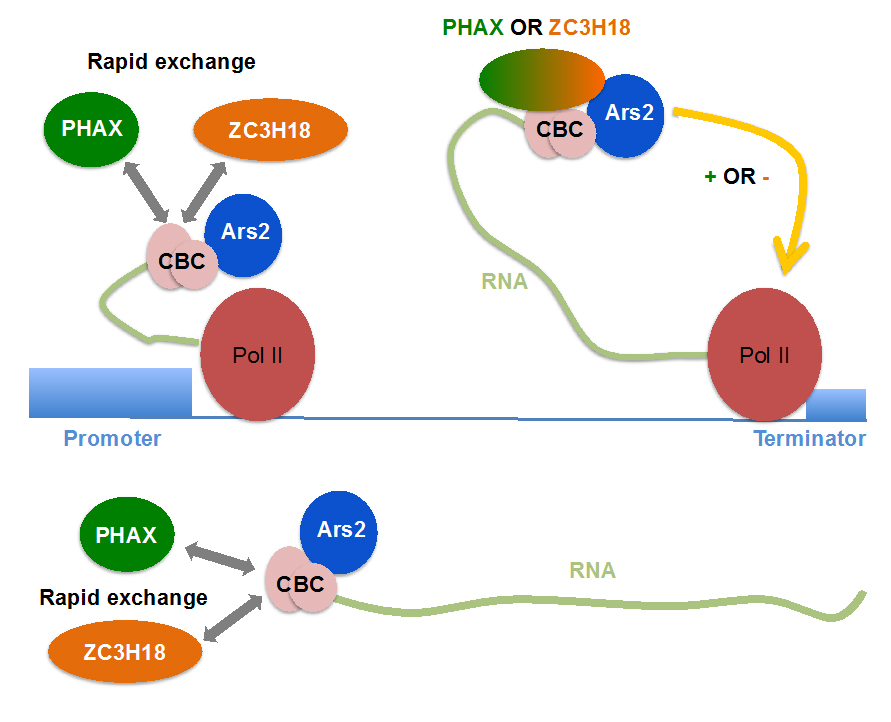
RNAs destined for degradation are diverse – short and long, immature and mature - so how are they identified? Well, a common feature for the large part of RNAs produced by the enzyme RNA polymerase II (PolII) is that their beginnings (their so-called ‘caps’) are bound be a protein complex called the ‘cap-binding complex’ (CBC) (see Figure). New findings, spearheaded by PhD student Simone Giacometti, working collaboratively between Aarhus, Montpellier and Edinburgh research institutions, now suggest that the CBC plays an important role, acting as a binding platform for additional proteins with roles in either RNA functionalization or degradation.
It was already known that the CBC is contacted by proteins important for RNA function (e.g. the PHAX protein, see Figure) and RNA degradation (e.g. the ZC3H18 protein, see Figure). Using a high throughput protein-RNA in vivo interaction method and measurements of the half-lives of relevant protein complexes, a model emerged that the PHAX and ZC3H18 proteins rapidly exchange on the CBC, initially offering the opportunity for both RNA production and destruction. The final decision about eventual RNA fate is probably taken when PolII reaches specific DNA/RNA elements (e.g. a terminator, see Figure), which locks one of the two defining complexes. This event constitutes a true quality control checkpoint for RNA production.
New research is now attempting to further test the validity of the proposed model as well as to identify additional proteins and protein complexes involved in the process. These findings are central for our understanding of how genomes work and how their massive RNA production is kept under control to avoid cellular chaos..
For further information, please contact
Professor Torben Heick Jensen
Department of Molecular Biology and Genetics
Aarhus University, Denmark
thj@mbg.au.dk; +45 60202705.
