Chain reaction in the human immune system trapped in crystals
A research team from Aarhus University has revealed details of how a chain reaction in the human immune system starts. With these results, the researchers hope to promote the development of strategies aimed at alleviating suffering caused by unintentional activation of the immune system.
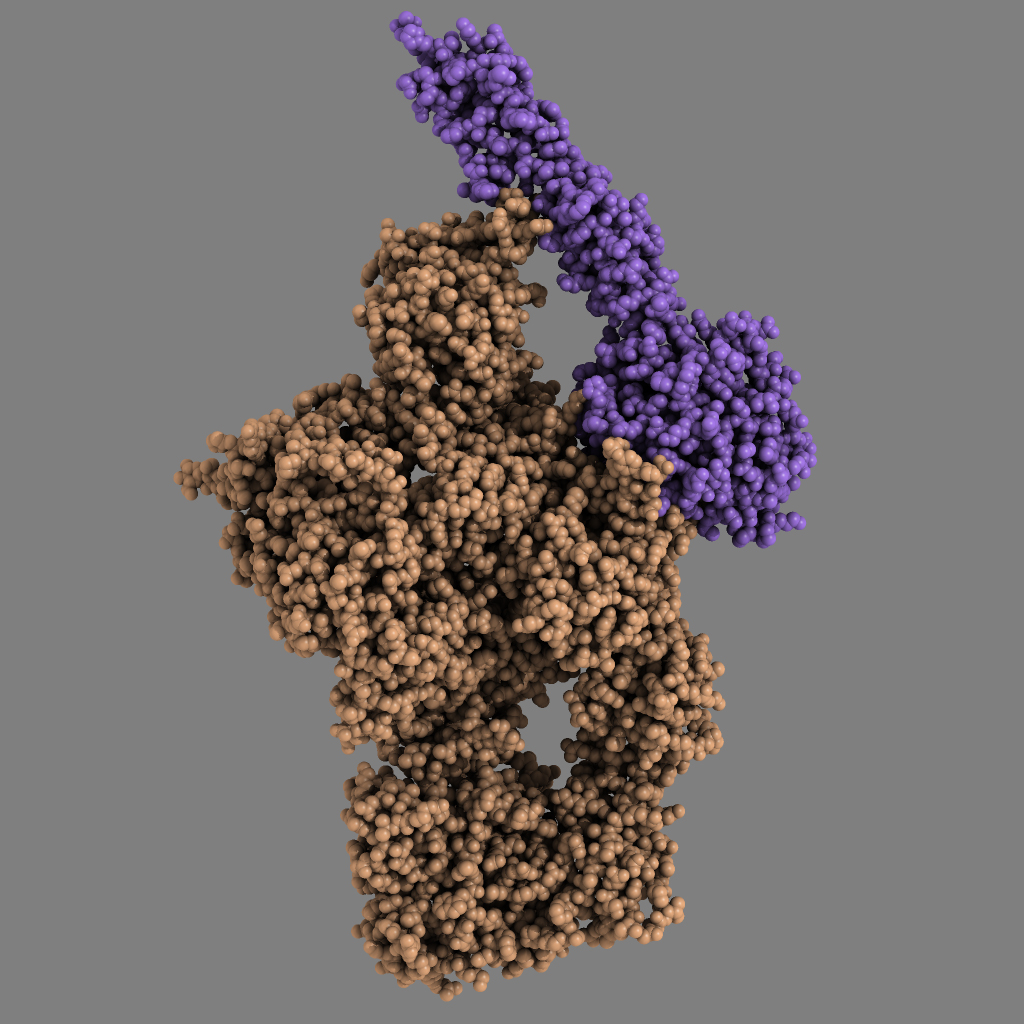
The complement system is part of the innate immune system and is composed of about 40 different proteins that work together to defend the body against disease-causing microorganisms. The complement system perceives danger signals in the body by recognising characteristic molecular patterns presented by pathogenic microorganisms or some of our own sick or dying cells that must be eliminated.
The complement system can be found in the blood, but also in the fluid surrounding the cells in tissues. Complement serves as the first line of defence against many pathogenic organisms, and its recognition of danger signals is handled by specialised proteins in the complement system.
For more than 20 years, Professor Steffen Thiel at the Department of Biomedicine, Aarhus University, has been a world leader in studies of MBL and MASP-2, which are two key proteins in the complement system. When MBL recognises the danger signal, MASP-2 is converted into an active enzyme that can now cleave the protein C4, a third important protein in the complement system. This cleavage is the first step in a chain reaction that ends with the elimination of pathogenic bacteria and dying cells. A research team led by Associate Professor Gregers R. Andersen at the Department of Molecular Biology and Genetics, Aarhus University, has now – in collaboration with Professor Thiel – succeeded in determining in atomic detail how the active enzyme MASP-2 recognises the substrate C4.
Crystals shot with X-rays
Two PhD students in molecular biology, Rune T. Kidmose and Nick S. Laursen, isolated the protein C4 from blood plasma, whereas they obtained the MASP-2 from Hungarian colleagues. They then crystallised the substrate C4 alone, but – to their surprise – they could also crystallise the enzyme-substrate complex C4·MASP-2. By exposing the resulting crystals to intense X-ray radiation, the two students managed within a year to determine the atomic structures of the C4 protein and the C4·MASP-2 complex. Two other PhD students, Sofia Sirotkina and Troels R. Kjaer, then performed laboratory experiments showing that the conclusions based on crystal structures were also valid when MASP-2 cleaved C4 in a test tube.
Potential impact on drug development
The results obtained by Rune T. Kidmose and Nick S. Laursen are remarkable. “It’s extremely rare that you can trap a proteolytic (protein-degrading) enzyme in the middle of the process of cleaving an intact protein,” says Associate Professor Gregers Rom Andersen, the students’ supervisor. “We now know in detail which parts of MASP-2 recognise the substrate C4. Another fascinating aspect of their results is that we also know the structure of both C4 and MASP-2 alone, so we can see how both the enzyme and the substrate change their three-dimensional structure when C4 is recognised by MASP-2. We can also see how these changes directly contribute to facilitating the cleavage of C4,” he concludes.
For Professor Thiel, the new results represent a completely new way to visualise how MASP-2 – whose function he discovered in 1997 – carries out its function. “It’s also a great personal pleasure to be involved right from the discovery of a new protein to the point where you obtain knowledge at the atomic level on how this protein functions as an enzyme,” he says.
In many situations, an undesirable activation of the complement system takes place, which can damage our own tissues. Several pharmaceutical companies are currently working on developing drugs that can attenuate such damage. “The very detailed understanding that we now have concerning parts of the complement system will undoubtedly lead to more intelligent ways of developing new drugs,” says Professor Thiel. “Of course, we won’t stop our studies here, as we’ll continue the very detailed studies of other proteins and molecular mechanisms within the immune system,” he continues.
The new results from Aarhus University have just been published in the prestigious journal Proceedings of the National Academy of Sciences (PNAS).
Rune T. Kidmosea, Nick S. Laursena, József Dobób, Troels R. Kjaerc, Sofia Sirotkinaa, Laure Yatimea, Lars Sottrup-Jensena, Steffen Thielc, Péter Gálb, and Gregers R. Andersena
Departments of aMolecular Biology and Genetics and cBiomedicine, Aarhus University, DK-8000 Aarhus, Denmark; and bInstitute of Enzymology, Research Centre for Natural Sciences, Hungarian Academy of Sciences, H-1113, Budapest, Hungary
More information
Associate Professor Gregers Rom Andersen
Department of Molecular Biology and Genetics
Aarhus University
Denmark
gra@mb.au.dk – +45 871 55507
Professor Steffen Thiel
Department of Biomedicine
Aarhus University
Denmark
st@microbiology.au.dk – +45 2927 0890
Translation: Lisbeth Heilesen
