A newly developed RNA molecule binds to SARS-CoV-2 virus and blocks its entry into cells
A research team from MBG and iNANO at Aarhus University has used an in vitro selection technique to develop an RNA molecule that recognizes and binds the Spike protein from SARS-CoV-2 with very high specificity and strength. The hope is that the new molecule, known as an RNA aptamer, can help improve diagnostics and treatment of COVID-19.
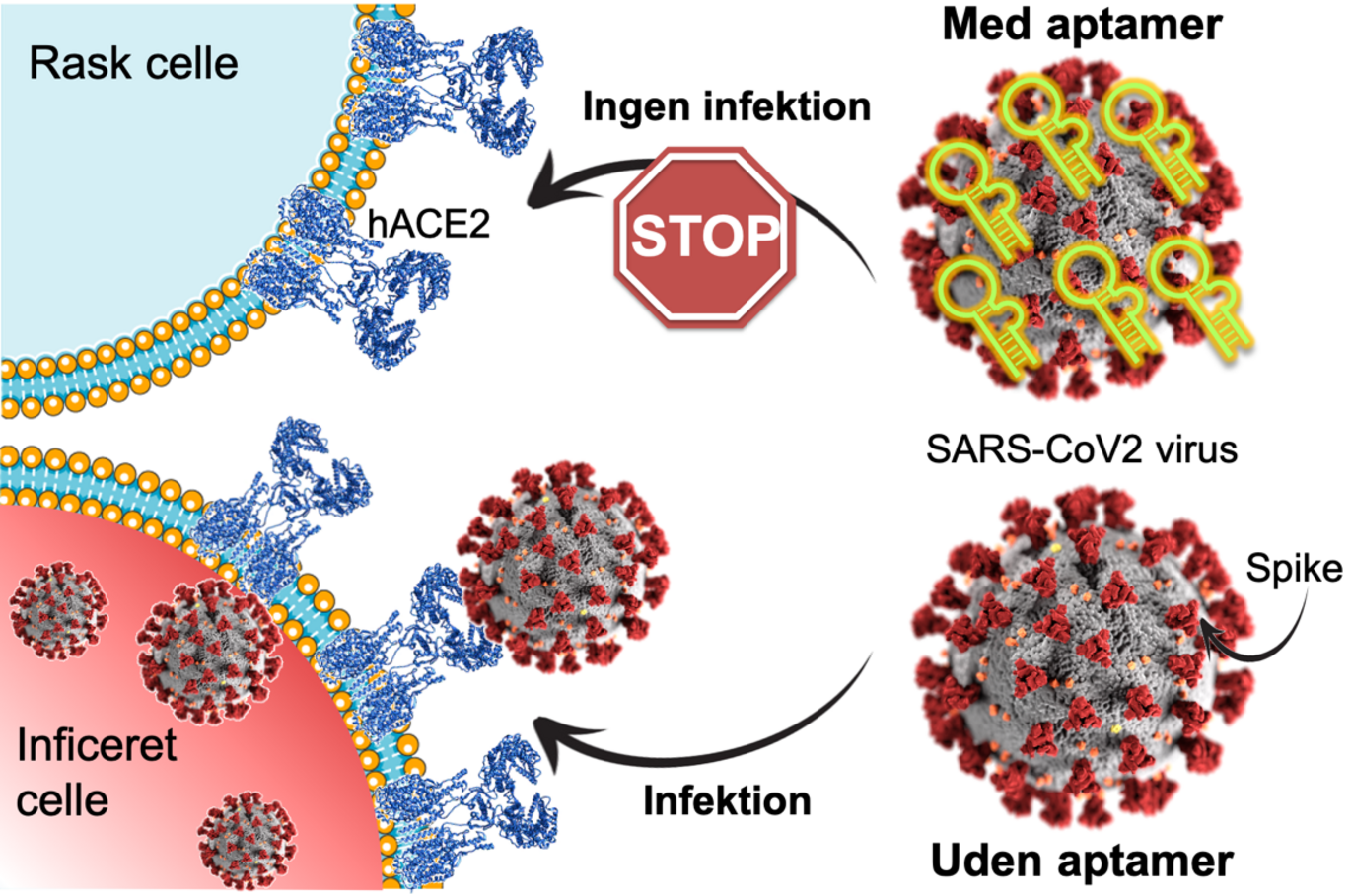
The past year and a half of COVID-19 pandemic has highlighted the importance of developing new molecules to diagnose and treat viral diseases. A new research article in PNAS from Julián Valero, Laia Civit, Jørgen Kjems and colleagues in Aarhus and in Regensburg in Germany describes the development of an RNA aptamer with very high binding affinity for the Spike protein that protrudes from the surface of SARS-CoV-2 virus particles. By attaching itself to the virus surface, the RNA aptamer prevents the Spike protein from serving as a key that allows the virus to enter a cell. The RNA aptamer folds into a 3D structure that makes it possible to recognize a specific target molecule of interest. In this way, aptamers can be compared to traditional antibodies, but they are much easier and cheaper to manufacture.
The new study shows that the aptamer efficiently blocks the binding between the Spike protein on the virus and the receptor ACE2 on human cells, thereby neutralising the infection of human cells grown in culture. The study also shows that the RNA aptamer inhibits the first known variants of the virus with comparable efficiency. “Since then, we have continued our studies and been able to show that it also recognizes the delta variant. We are now waiting for samples of the newly identified variant, omicron, so we can test whether the aptamer also recognizes that, ”says Jørgen Kjems.
The promising results point towards a possible future use of the aptamer in COVID-19 treatment and towards the development of cheaper and more sensitive diagnostic tests. Julian Valero, the first author on the paper, also highlights the applications beyond COVID-19: 'We can apply this technology to rapidly develop new RNA aptamer molecules targeting emerging SARS-CoV-2 variants or other viral threats'.
The research project is supported by the Carlsberg Foundation and by the EU FET-open project VIROFIGHT. Jørgen Kjems and Julián Valero are both affiliated with CellPAT, a center of excellence housed at iNANO and funded by the Danish National Research Foundation.
For more information, please contact
Professor Jørgen Kjems, Department of Molecular Biology and Genetics/Interdisciplinary Nanoscience Center (iNANO), Aarhus Universitet, jk@mbg.au.dk, 2899 2086
Adjunkt Julián Valero, Interdisciplinary Nanoscience Center (iNANO), Aarhus Universitet, jvalero@inano.au.dk
External financing: Carlsbergfondet bevillingsnr. CF20-0045, Danmarks Grundforskningsfond bevillingsnr. DNRF135, EU Horizon2020 bevillingsnr. 899619
Conflicts of interest: None
Other: Aarhus University has submitted a patent application for the developed RNA aptamer
Valero J*, Civit L,* Dupont DM, Selnihhin D, Reinert LS, Idorn M, Israels BA, Bednarz AM, Bus C, Asbach B, Peterhoff D, Pedersen FS, Birkedal V, Wagner R, Paludan SR, Kjems J: A serum-stable RNA aptamer specific for SARS-CoV-2 neutralizes viral entry. PNAS December 14, 2021 118 (50) e2112942118; doi.org/10.1073/pnas.2112942118.
