How to keep the nucleus clean
RNA turnover in the nuclei of eukaryotic cells is controlled by the RNA exosome aided by numerous cofactors. Researchers at Aarhus University and Copenhagen University now show how two major nuclear exosome cofactors recognize their RNA targets to keep a clean nuclear environment. This is important for the health of our cells – and thus humans.
Cells are small factories that constantly produce protein and RNA molecules by decoding the genetic information stored in the DNA of their chromosomes. The first phase of this decoding, the transcription process, ”transcribes” the DNA code into RNA molecules. In humans, and most other organisms, all cells of the body carry the full genetic information of the entire organism, with each individual cell requiring only a small subset of its DNA decoded. Even so, the first decoding phase (transcription) is pervasive and produces a large amount of surplus RNAs.
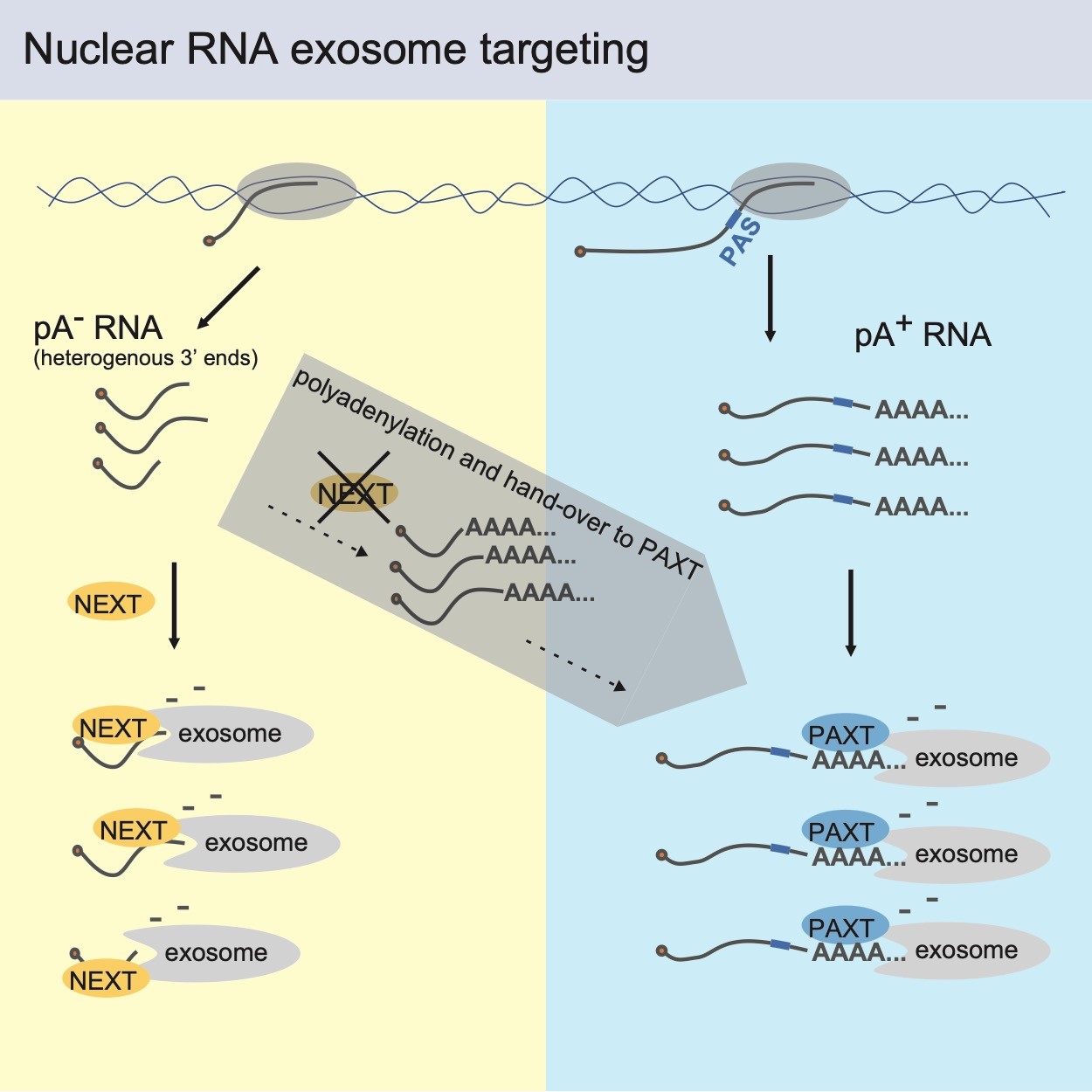
These extraneous RNAs do, however, not accumulate, as they are degraded shortly after their production. This prevents deleterious accumulation of non-functional transcripts that would otherwise be detrimental to cell health. Most of this RNA decay is carried out by the nuclear RNA exosome complex, an RNA 3’-5’ exonuclease, which is recruited to RNA by specific adapters, like the so-called NEXT complex and the PAXT connection. The laboratory of Torben Heick Jensen previously found that NEXT and PAXT aid in decay of different kinds of nuclear RNAs, but how specificity is achieved remained enigmatic.
With their new publication, the research team now reveals that NEXT substrates contain ‘naked’ 3’ends, whereas PAXT substrates harbor so-called poly(A)-tailed 3’ends. Despite this clear division, the study also reveal that the decay systems can co-operate, which helps cells to degrade NEXT substrates, even in the potentially hazardous situation when NEXT activity is decreased. In this case, NEXT targets (which are normally produced without a poly(A) tail and swiftly removed) acquire poly(A) tails – a hallmark of PAXT targets – which subject them to PAXT-mediated decay. In conjunction, this provides a two-layered targeting mechanism for the efficient nuclear sorting of the human transcriptome. An important question now remaining to be answered is how this efficient sorting is coupled to the production (transcription) of RNA.
The findings result from a collaborative project spearheaded by postdoc Guifen Wu and team leader Manfred Schmid from the laboratory of Torben Heick Jensen at the Department of Molecular Biology and Genetics, Aarhus University, in collaboration with Leonor Rib and Albin Sandelin from the Department of Biology and Biotech Research and Innovation Centre, University of Copenhagen. The study was financed by the Novo Nordisk Foundation and the Lundbeck Foundation, and published in the international journal Cell Reports.
‘A two-layered targeting mechanism underlies nuclear RNA sorting by the human exosome’ by Guifen Wu, Manfred Schmid, Leonor Rib, Patrik Polak, Nicola Meola, Albin Sandelin, Torben Heick Jensen. Cell Reports.
For further information, please contact
Postdoc Guifen Wu- wuguifen@mbg.au.dk
Teamleder Manfred Schmid- ms@mbg.au.dk
Professor Torben Heick Jensen - thj@mbg.au.dk – mobile: +45 60202705
Department of Molecular Biology and Genetics, Aarhus University, Denmark
