New research shows how important protein keeps our cell membranes in balance
In a comprehensive study published in the journal Nature Communications, researchers have studied the structure, function and mechanisms of the protein ATP8B1, which flips lipid molecules in our cell membranes and plays a key role in bile biosynthesis. Importantly, ATP8B1 has also emerged as a genetic marker of Alzheimer’s resilience. These results give new possibilities for therapeutic interventions in conditions related to bile homeostasis disorders and neurodegenerative diseases.
Lipids are the main constituents of our cell membranes, which are formed as lipid bilayers. The distribution of lipids is far from uniform; it is asymmetric, with different lipid compositions in the outside and inside layers. This asymmetry is essential for a variety of cellular functions, from maintaining membrane homeostasis to enabling cell signaling and numerous other physiological processes at or across membranes.
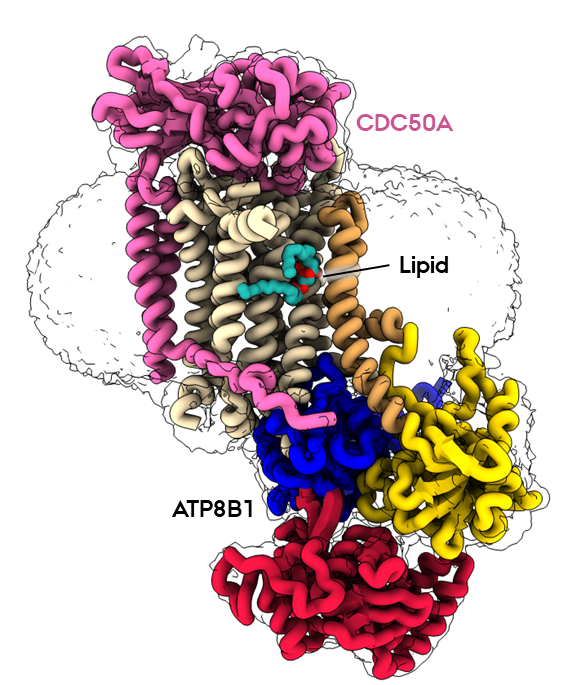
P4-ATPases, also known as flippases, are key players in creating and maintaining this lipid asymmetry. These enzymes actively transport lipids from the outside (exoplasmic) leaflet to the inside (cytosolic) leaflet coupled to ATP hydrolysis and ensure the proper distributions of lipids. The ATP8B1-CDC50A flippase complex in particular, has been the subject of the current study, leading to several new, groundbreaking discoveries.
The function of ATP8B1 lipid flippase is critical for the regulation of bile production, a vital substance in our digestive system, but the direct link within bile producing liver cells remains unknown. Additionally, recent studies have spotlighted the relevance of genetic variants in the regulatory segment of the ATP8B1 gene as a strong genetic marker for Alzheimer's resilience. Interestingly, mutations that impair function of the closely related ATP8B4 lipid flippase are oppositely important risk factors of Alzheimer’s. It is therefore of strong interest to understand how ATP8B1 is linked to these processes and pathologies.
In the new study, the research team employed state-of-the-art cryo-electron microscopy techniques to capture nine different states associated with the lipid transport and determine structures at 2.4 to 3.1 Å resolution for these states. These structural insights, combined with functional and computational studies, reveal the inner workings of the human flippase ATP8B1-CDC50A complex and its fine regulation by specific regulatory lipids known as phosphoinositides, or PIPs.
These findings open doors to a deeper understanding of how lipid flippases operate and the intricate roles they play in cellular processes and are regulated. Importantly, the study also resolves earlier discrepancies about the ATP8B1 transport substrates.
Noteworthy, both up- or down-regulation of ATP8B1-CDC50 function could potentially be of interest in drug discovery, and the study also reveals critical information to such applications.
The team behind the new results in Nature Communications is a collaboration between researchers from Aarhus University (DANDRITE, Dept. Molecular Biology and Genetics, and iNANO), the University of Copenhagen, Oxford University, and the University of Paris-Saclay.
The results were published in the international journal Nature Communications
For further information, please contact
Professor Poul Nissen
Department of Molecular Biology and Genetics & DANDRITE
Aarhus University, Denmark
Email: pn@mbg.au.dk. mobile: +4528992295
SUPPLEMENTARY INFORMATION
We strive to ensure that all our articles live up to the Danish universities' principles for good research communication. Against this background, the article is supplemented with the following information:
Study tipe | Experiment |
External funding | This work was supported by a Marie Skłodowska-Curie Actions Individual Fellowship — 590 LivFlip (101024542) to T.D., by Engineering and Physical Sciences Research Council grants 591 (EP/X035603 and EP/V030779) to S. K., by an ANR grant (ANR-14-CE09-0022) to G.L., , made available under aCC-BY-NC-ND 4.0 International license. (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is bioRxiv preprint doi: doi.org/10.1101/2023.04.12.536557; this version posted May 4, 2023. The copyright holder for this preprint 20 592 by the French Infrastructure for Integrated Structural Biology (FRISBI; ANR-10-INSB-05) to 593 G.L., by the Centre National de la Recherche Scientifique (CNRS) to G.L., by the Lundbeck 594 Foundation to the BRAINSTRUC structural biology initiative (155-2015-2666) to K.L.-L. and 595 P.N., and by a Lundbeckfonden Professorship grant (R310-2018-3713) to P.N |
Conflicts of interest | None |
Link to scientific paper | Thibaud Dieudonné, Felix Kümmerer, Michelle Juknaviciute Laursen, Charlott Stock, Rasmus Kock Flygaard, Syma Khalid, Guillaume Lenoir, Joseph A. Lyons, Kresten Lindorff-Larsen and Poul Nissen. DANDRITE, Nordic EMBL Partnership for Molecular Medicine, Department of Molecular Biology and Genetics, Aarhus University, Aarhus, Denmark Structural Biology and NMR Laboratory & Linderstrøm-Lang Centre for Protein Science, Department of Biology, University of Copenhagen, Denmark Department of Biochemistry, University of Oxford, Oxford, United Kingdom Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC),91198, Gif-sur-Yvette, France Department of Molecular Biology and Genetics, Aarhus University, Aarhus, Denmark “Activation and substrate specificity of 1 the human P4-ATPase ATP8B1” Nature Communications |
