New knowledge may reduce stroke-related injuries
[Translate to English:] Danske forskere finder vigtigt nyt om fællestræk ved udvikling af Alzheimers sygdom, type II diabetes, maniodepressive lidelser og celledød efter hjerneblødning, der kan muliggøre en tidlig forebyggelse og behandling.
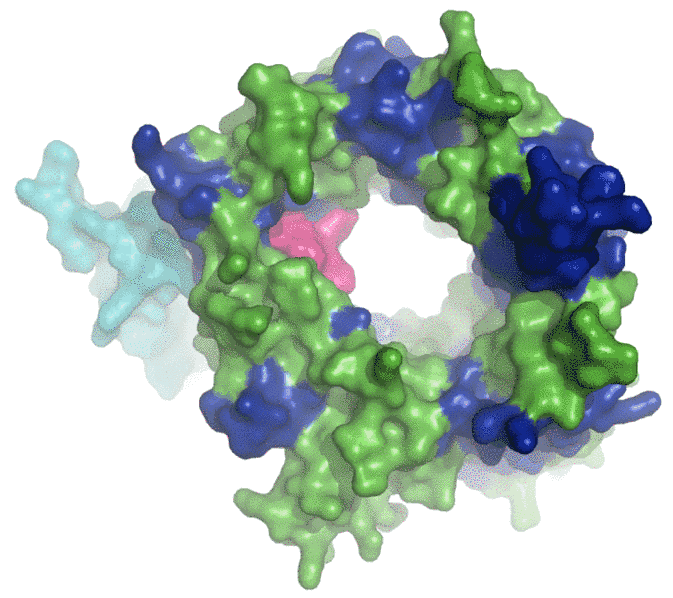
Researchers at the Lundbeck Foundation's MIND Center at Aarhus University publish the three-dimensional atomic structure of the protein sortilin in the journal Nature Structural and Molecular Biology. Sortilin contributes to cell death in the brain after bleeding and blood clots. The new knowledge about the structure gives a deeper understanding of some basic processes that are active in cell death in the brain, in developing Alzheimer's disease, type II diabetes and manic-depressive disorders. Knowledge of sortilin's structure will make it possible to design a molecular marker that makes it easier to see the difference between normal and diseased cells, thus improving the prospects for early preventive or therapeutic intervention.
The determination of sortilin's structure is a result of the long-standing collaboration between research groups at the Departments of Medical Biochemistry and Molecular Biology at Aarhus University, headed by Professor Claus Munck Petersen and Associate Professor Søren Thirup, respectively.
The discovery of the sortilin protein
Sortilin was first isolated in 1997 at the Department of Medical Biochemistry, Aarhus University, where in 2004 it was also shown that under certain circumstances sortilin causes brain cells to die. It was later discovered that sortilin is one of a family of five closely related proteins that are involved in such diseases as Alzheimer's, type II diabetes and manic-depressive disorders.
Sortilin is mainly found in brain tissue, where it binds to the surface of the cells and can bind to other proteins (ligands). One of these ligands can - when it binds to sortilin - get the cell to commit suicide, which is what happens with blood clotting and bleeding in the brain.
Several very different ligands to sortilin have been identified, and so far it has been a mystery why none of them can bind to sortilin at the same time. However it is obvious now that the structure has been determined. Sortilin turns out to be the largest so-called propeller structure ever seen (see Figure), and this leaves room for a central tunnel where each ligand can be recognized specifically. But because it is a tunnel, there is only room for one ligand at a time.
Design of new markers
With this new structure the researchers hope to be able to design a ligand to sortilin that will prevent other ligands from binding and thus prevent the brain cells from committing suicide. Although it is reasonable to assume that the same mechanism occurs in Alzheimer's disease this is not the case. In Alzheimer's we are dealing with the protein APP, which - for lack of protein SorLA (member of the sortilin family) - is degraded wrongly and causes damage in the brain. To solve this problem the researchers hope to develop a marker which will enable them to provide an early diagnosis by brain imaging.
Further information
For further information please contact Claus Munck Petersen, Department of Medical Biochemistry, Aarhus University (cmp@biokemi.au.dk) cell no. +45 8942 2865 or Soren Thirup, Department of Molecular Biology, Aarhus University (sth@mb.au.dk) cell no. +45 6110 9126.
Figure (of the protein) in high resolution
Link to the article publiced in Nature Structural and Molecular Biology:
Ligands bind to Sortilin in the tunnel of a ten-bladed ![]() -propeller domain
-propeller domain
Esben M Quistgaard1, Peder Madsen2, Morten K Grøftehauge1, Poul Nissen1, Claus M Petersen2 & Søren S Thirup1
- MIND Centre, Department of Molecular Biology, University of Aarhus, Gustav Wieds Vej 10C, DK 8000 Århus C, Denmark.
- MIND Centre, Institute of Medical Biochemistry, University of Aarhus, Ole Worms Allé 1170, DK 8000 Århus C, Denmark.
