Seeing is believing. We use zebrafish as a model organism to "visualize" unsolved mysteries in biology – the role of extracellular vesicles (EVs) in health and disease. EVs emerge as a new frontier that has the potential to redefine today's knowledge across many research areas from basic to clinical sciences. Our lab is where nanoscience meets zebrafish in search of new therapeutic inspirations by learning, manipulating and mimicking nature's smart biomolecular architecture, EVs.
Our current interest centres around EV biology, for which we have just started a journey to explore their role as a cell-to-cell messenger in tissue injury, inflammation and regeneration. Why? It's because EVs can be therapeutic targets for molecular intervention or potentially mimicked as "on-demand" drug delivery vehicles. However, much is still not known about these natural nanoparticles that carry biomolecular cargoes. For example, by which mechanisms can EVs find the target recipient cells via the bloodstream? What kind of messages are conveyed to regulate/support the recipient cells? With zebrafish as our little partners, we seek answers to these questions by nanoscience approaches, genetic engineering, bioinformatics and live imaging of transgenic embryos.
We do both basic and applied sciences. Apart from the fish, we strongly collaborate with nanoscientists at iNANO and health science researchers at the Department of Biomedicine and the University Hospital to make our interdisciplinary research happen!
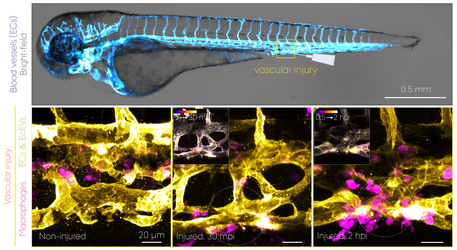
But why zebrafish, not mice? We can manipulate their genome e.g. to label cells of interest by fluorescent (and functional) proteins, and it allows us to image live embryos in real-time seeing through the tissues non-invasively (click the "Our Bioimaging Strategy" button below to expand). A particularly successful example of our approach is live imaging and electron microscopy of how nanoparticles injected into the bloodstream are captured by macrophages – the cells that eat/clean up foreign materials, pathogens and dead cells just as they do in mice and humans. Therefore by looking at the biology evolutionarily conserved at the cellular and molecular levels, we can study how life behaves at the interface with nanotechnology.
Transgenic lines with a cell type-specific fluorescent protein reporter allow us to study the dynamic behaviour of cells such as macrophages and how (injected) nanoparticles are cleared from the bloodstream in a living organism. This intravital whole-embryo bioimaging approach can also be combined with a high-resolution modality, transmission electron microscopy (TEM), to support the real-time observations by visualizing those biological processes at the nanoscale. The unrivalled strength of the zebrafish model is not only for bioimaging itself, but also its screening capacity that enables testing of multiple nanoformulations within a short time period. Shown above is a summary of results obtained using double transgenic zebrafish embryos intravenously injected with 70 nm SiO2 nanoparticles. dpf, days post-fertilization. mpi/hpi, minutes/hours post-injection. (Image: Yuya Hayashi. Adapted from Hayashi et al. (2020) & Mohammad-Beigi et al. (2020) ACS Nano. Copyright 2020 American Chemical Society)
Macrophages (magenta) with internalized nanoparticles (cyan) crawling along the inner side of blood vessels (grey). Tg(fli1a:EGFP); Tg(mpeg1:mCherry) embryos at 3 dpf were injected with Pacific Blue-labelled 70 nm SiO2 nanoparticles (2 ng). Time-lapse imaging was performed at the intervals of every 16 s for 15 min at 1-4 hpi. (Adapted from Hayashi et al. (2020) ACS Nano. Copyright 2020 American Chemical Society)
EVs secreted from the yolk syncytial layer (cyan) circulate in the bloodstream and are captured by endothelial cells (grey) and macrophages (magenta). Eggs obtained from Tg(kdrl:Hsa.HRAS-mCherry) or Tg(mpeg1:mCherry) fish were transfected with plasmid DNA by microinjection into the yolk syncytial later that is formed at 3-4 hpf. The caudal vein tissue of the transfected embryos at 3 dpf was imaged every minute for 30 min. (Movie: Yuya Hayashi)
"Differential Nanoparticle Sequestration by Macrophages and Scavenger Endothelial Cells Visualized in Vivo in Real-Time and at Ultrastructural Resolution" by Yuya Hayashi*, Masanari Takamiya, Pia Bomholt Jensen, Isaac Ojea-Jiménez, Hélicia Claude, Claude Antony, Kasper Kjær-Sørensen, Clemens Grabher, Thomas Boesen, Douglas Gilliland, Claus Oxvig, Uwe Strähle, and Carsten Weiss.
ACS Nano 14 (2020) pp. 1665-1681. https://doi.org/10.1021/acsnano.9b07233.
Exosomes: Decrypting "blood-streamed" RNA communication
From an organ to another organ, cells send signals to coordinate the physiology of the entire body. A well-known example is signalling by hormones, but what if cells instead wish to deliver more complex messages than signals? A striking discovery over the past decades is the packaged delivery of small RNAs in nano-sized vesicles called exosomes to "stream" the RNA language through the blood. Using zebrafish as an in vivo model, we study exosomes and other EVs of various origins on their dynamic behaviour in the bloodstream and how they are trapped by cells. We have also developed a method to profile the biomolecular cargo of EVs "in transit" from donor cells. Our aim is to capture snapshots of conversation between cells and thus to identify novel therapeutic targets, in particular those involved in tissue injury, inflammation and regeneration.
Our main collaboration partners for this project are Omiics ApS, Prof. Jørgen Kjems (iNANO, Aarhus University), Dr. Guillaume van Niel (INSERM, France), and Dr. Frederik Verweij (Utrecht University, the Netherlands).
What the Cell "Sees" in Extracellular Vesicle Biology
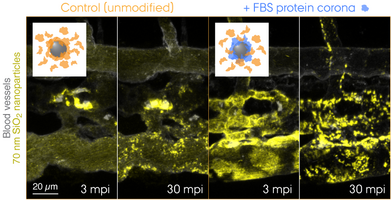
What lies at the interface of biological receptors and nanoparticles? Over the past decade, nanoscientists have studied complex biophysical interactions that take place between biomolecules and nanoparticles. Proteins are among those biomolecules that form a "corona" around the nanoparticle exhibiting a biologically meaningful residence time at the surface – long enough for triggering e.g. receptor-mediated endocytosis. Today, it is widely accepted that the cell "sees" the surrounding proteins, not the bare surface of the nanoparticle. Now, the question is whether this holds true for EVs, or should we rather say, biological nanoparticles. The biomolecular corona field is gradually merging with the emerging EV research, and there is an urgent need for technology development to tackle this challenging problem. Combining EV engineering and proteomic approaches, we aim to redefine EV biology with a new dimension, fundamentally from the cell's viewpoint.
"Tracing the In Vivo Fate of Nanoparticles with a 'Non-Self' Biological Identity" by Hossein Mohammad-Beigi, Carsten Scavenius, Pia Bomholt Jensen, Kasper Kjær-Sørensen, Claus Oxvig, Thomas Boesen, Jan J. Enghild, Duncan S. Sutherland, and Yuya Hayashi*.
ACS Nano 14 (2020) pp. 10666–10679. https://doi.org/10.1021/acsnano.0c05178.
Outwitting antiviral defence mechanisms
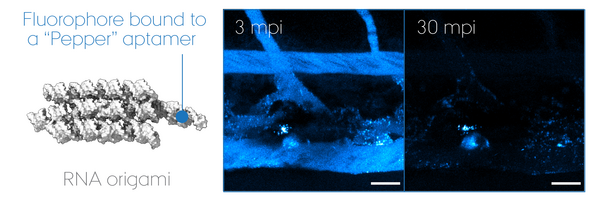
"RNA Origami" uses RNA as molecular building blocks that self-assemble into programmable 3D nanostructures. Today, much remains unanswered about its potential use for therapeutics, despite that non-self nucleic acid components such as those of virus are inherently immunogenic. We focus on its biomedical application using zebrafish embryos as a screening model to develop a multi-functional RNA origami without undesired antiviral responses. In particular, we look at key determinants of nucleic acid sensing and inflammation to unravel how life "sees" artificial RNA architectures and thus how we can outsmart it to accomplish intended functionalities in a living organism. Our goal is to pave the road for a safe-by-design approach in the RNA nanotechnology field and ultimately accomplish mimicry of extracellular RNA communication powered by EVs.
RNA origami is a nascent technology invented by Dr. Ebbe Sloth Andersen and Prof. Jørgen Kjems (iNANO, Aarhus University). Together with a new PI on nanomedicine, Dr. Julián Valero Moreno, they are important collaborators on the designing part of RNA nanotechnology.
Dr. Jean-Pierre Levraud (NeuroPSI & Institut Pasteur, France) as well as Dr. David Olagnier and Prof. Søren Riis Paludan (Department of Biomedicine, Aarhus University) are collaboration partners on antiviral immunity in zebrafish.
Bachelor, Master, medical and visiting (Erasmus+) students are always welcome to contact Yuya (yuya.hayashi@mbg.au.dk) and he will usually respond very quickly ;) Student projects are designed according to your scientific interests (well, motivation is the key driving factor to complete a project!) but we also help you to define the project based on what we can offer. Once the project starts, you will become a group member and report to weekly group meetings.
On the Group Members page, you can have an idea about what kind of projects each member works on.
Listed below are the scientifc topics that we cover, and techniques that you can learn from us.
Will you be writing a thesis (Bachelor's/Master's)? We understand that it is a scary thing that concludes an academic degree after exposing yourself to broad knowledge through many courses. As a project for the University Pedagogical Course, Yuya dedicated his time in setting up a pseudo-course in Brightspace to which the group members are all invited. In this technology-enhanced learning platform, students can check the thesis writing schedule, watch video lectures by Yuya on how to write a thesis, and participate in guided peer feedback activities. The platform will be opened every semester, and continue to be updated in response to students' need. So, if you consider yourself as one who wants a lot of help during the stressful period of thesis writing, Yuya's supervision style will suit you nicely ;)
We use Slack for quick communication wiwthin the group. In this way, Yuya prioritizes helping students in the lab who have everyday questions and reporting. Slack is also used to facilitate other thesis writing activities outside Brightspace. If you join the group, you'll never be left alone not knowing what to do!


We are very proud of having a stereomicroscope with an integrated camera for easy and simultaneous monitoring of the specimen on a display. Students quickly learn about the different developmental stages of zebrafish embryos and how they are handled under the microscope. It has proved especially useful for training of microinjections and needle-wounding. Snapshots and video recording can also be done by a remote controller, if students observe something unusual or worthy reporting to the supervisors ;)
Not only project supervision, we also contribute to lectures, theoretical exercises and laboratory practicals in courses offerred across the university.


Yuya is a founder & organizer of the AU local network for extracellular vesicle research, EVAnet. The EVAnet Student Meeting series is arranged for students at all levels from bachelor to PhD and postdocs to facilitate discussion forums and networking activities in a relaxed atmosphere. Students who work on topics related to extracellular vesicles are all encouraged (not obliged!) to participate, not just for learning, but also to get to know people in the same research field ;)


We use zebrafish embryos until 120 hpf. While students can carry out short-term experiments within this period, access to the fish facility requires specific training for handling animals in research.
Our primary workhorse for bioimaging and screening of zebrafish embryos is a Leica THUNDER Imager Model Organism coupled with a microinjector (see here for details). For advanced imaging, we also do confocal laser scanning microscopy, light-sheet microscopy and (correlative light-) electron microscopy through collaborations and core facilities.
We have the standard tools for routine PCR and electrophoresis of agarose gels and SDS-PAGE & Western blots. Bacterial, cell culture and qPCR work are carried out in Prof. Claus Oxvig's lab next doors.
We have access to the research infrastructure at iNANO for materials characterization.
The local EV research network "EVAnet" facilitates the sharing of expertise and research infrastructrue across the university.

The zebrafish facility here at MBG has assisted our research since 2014. A number of genetically modified lines generated by us or other zebrafish scientists are available at the facility. Among those, we routinely use those with a fluorescent reporter transgene that labels cells of particular interest for e.g. live imaging. Generation and establishment of a new transgenic line takes min. 7 months but normally longer than that. Enquiries regarding the use of the fish facility should be made to Dr. Kasper Kjær-Sørensen or Prof. Claus Oxvig.
Blood vessels (Endothelial cells)
Tg(fli1a:EGFP)y1 ZFIN
Tg(kdrl:Hsa.HRAS-mCherry)s896 *Membrane-labelling ZFIN
Embryonic macrophages
Tg(mpeg1:EGFP)gl22 ZFIN
Tg(mpeg1:mCherry)gl23 ZFIN
Tg(mpeg1:EGFP-Mmu.Rpl10a)Auzf9 *TRAP ZFIN
Neutrophils
Tg(lyz:DsRed2)nz50 ZFIN
Tumour necrosis factor-alpha (as a signature of macrophage activation)
Tg(tnfa:EGFP-F)ump5 *Membrane-labelling ZFIN

Currently, we are sharing the lab space with a new group led by Dr. Gilles Vanwalleghem, who is also a team leader at DANDRITE and an expert in bioimaging of zebrafish. We will have regular joint group meetings to exchange relevant skills and knowledge and discuss lab issues. Together with his group, we thrive to promote the zebrafish initiatives here at MBG ;)
Our neighbours are people from Prof. Claus Oxvig's, Dr. Bo Thomsen's, Dr. Lisbeth Schmidt Laursen's and Prof. Daan van Aalten's groups who work with cell cultures, fish and mouse models. Students, PhDs and postdoc researchers thus have a vibrant social life in a mixed Danish-international work environment.
We are constantly looking for students who wish to join the group and carry out a project as a part of the educational programme. Please contact Yuya to hear about project opportunities.
We have collaborations with health science researchers, for example, at Department of Biomedicine and Department of Clinical Medicine (Aarhus University Hospital). Medical students who are interested in our research are advised to contact me to find project opportunities through such collaborations.
ERASMUS students/trainees from abroad are always welcome to take a part in our research. Please send me an e-mail expressing what skills you wish to learn and we can discuss about projects for the mutual benefit! There are also possibilities of financial support for foreign students through an internal grant from the Department.
We currently have no openings.
Please contact Yuya Hayashi (yuya.hayashi@mbg.au.dk) to discuss possibilities.