A study from Torben Heick Jensen’s group at the Department of Molecular Biology and Genetics, Aarhus University, uncovers a new fail-safe mechanism for RNA decay that ensures that non-productive RNA is efficiently degraded in cells that experience external stress

Researchers have uncovered a surprising mechanism for how RNA polymerase II (RNAPII) transcription is terminated in eukaryotic cells. This involves simple DNA sequences; called T-tracts, which were known to terminate RNA polymerase III (RNAPIII) as well as prokaryotic RNA polymerase. The finding therefore suggests a broadly conserved role for T-tracts in finalizing transcription.

Work from Torben Heick Jensen’s research group at the Department of Molecular Biology and Genetics adds a new layer of complexity to selective RNA degradation. The researchers have discovered an adaptor complex, vNEXT, that recognises RNA structure rather than sequence when bringing target RNAs to the nuclear exosome.

In mammalian cells, RNA production often encounters obstacles during the early stages of transcription. In a special issue of Molecular Cell focussed on RNA biology, William Garland and Torben Heick Jensen review the early quality control mechanisms that shape the short RNA landscape in our cells.
Transcription is the process through which DNA is copied to RNA. Surprisingly, only a minority of transcription events reach completion, supposedly due to quality control of the transcription reaction. New results now add to the pathways by which this is achieved.
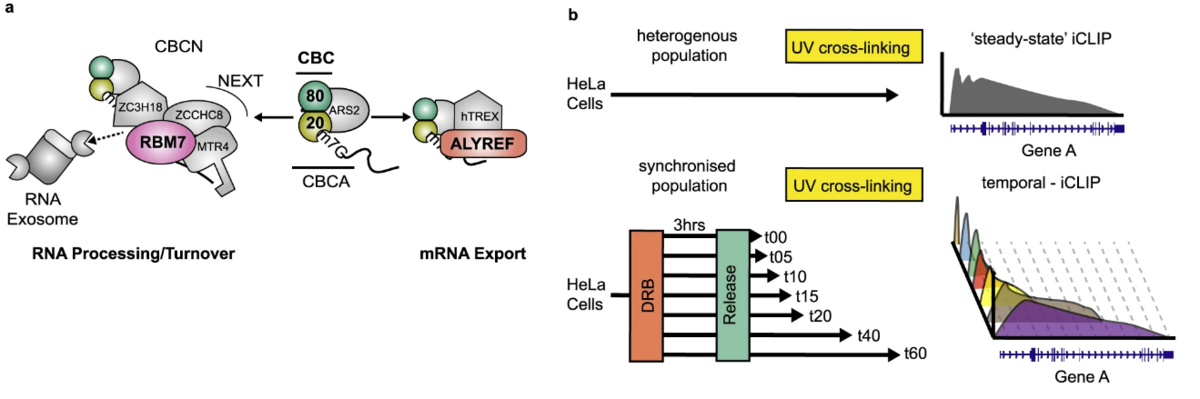
More than 80% of the human genome is transcribed, generating a large ensemble of nascent RNA that is co-transcriptionally packaged by RNA-binding proteins (RBPs). This vast genomic output raises an important question – where and when do RBPs associate to direct all of these transcripts into their appropriate processing/export or turnover pathways? Researchers from Denmark and Scotland now address this question in a new article in Nature Communications.
In a joint collaboration, researchers from Aarhus, Cambridge and Warsaw have characterized how the essential mRNA poly(A) tails are synthesized in the budding yeast Saccharomyces cerevisiae. The study reveals several mechanisms controlling poly(A) tail lengths and hereby ensuring the robustness of gene expression.
RNA turnover in the nuclei of eukaryotic cells is controlled by the RNA exosome aided by numerous cofactors. Researchers at Aarhus University and Copenhagen University now show how two major nuclear exosome cofactors recognize their RNA targets to keep a clean nuclear environment. This is important for the health of our cells – and thus humans.
Nuclear RNA levels are kept in check by RNA decay factors. Now, researchers at Aarhus and Copenhagen Universities show that an excess of RNA in the nucleus can have negative effects on a crucial regulator of stem cell differentiation.
How cells translate their genetic information into functional RNA and protein is a central question in biology. Researchers from the Department of Molecular Biology and Genetics at Aarhus University have invented a new technology to study regulatory principles of gene expression. Applying this methodology to bakers yeast they found that RNAs that fail to get exported from their site of synthesis in the cell nucleus are rapidly decayed. This provides a new regulatory principle in the complex process of gene expression.
Researchers at Aarhus University have characterized the human RNA decay factor ZC3H18 and discovered its unanticipated role in the production of protein-coding RNA. The new study, published this week in <em>Cell Reports</em>, therefore reveals a double-faced activity of ZC3H18 in nuclear RNA fate decisions.
Our genomes are promiscuously transcribed into RNA. How cells manage to sort this massive genomic output into functional and non-functional material has remained enigmatic. New research describes protein interactions involved in such RNA fate determination.
Genomes are promiscuously transcribed into RNA. However, not all of this material is immediately useful, which means it has to be targeted and degraded in order to sustain cellular life. A newly discovered RNA decay pathway functioning inside human nuclei does just that.
Intense investigations during the past 10-15 years have revealed that the human genome is transcribed in a manner that is much more complicated than previously appreciated. A collaboration between researchers from Aarhus and Copenhagen now reveals some underlying principles leading to such promiscuous genome activity.
A new important role for a protein connected to the proper function of neurons has been discovered by a research group from MBG, Aarhus University. The studies shed new light on gene expression regulation and may ultimately lead to an understanding of how neurological defects occur when this protein is mutated.
In collaboration with two other European groups, researchers from Aarhus University have uncovered molecular details leading to targetting of the major RNA decay machinery, the RNA exosome, to its nuclear RNA substrates. Studies can now be designed to address the role of this early nuclear RNA decay pathway in processes where rapid RNA decay may be critical, i.e. during embryonic development, cell differentiation and various stress conditions.
Professor Torben Heick Jensen, Department of Molecular Biology and Genetics, Aarhus University, receives DKK 60 million (Euro 8 million) from the Novo Nordisk Foundation's Challenge Programme to establish the research center 'Exo-Adapt', which will determine how our cells sort genetic information

At the meeting of members on 31 March 2016 in the Royal Danish Academy of Sciences and Letters, Professor Torben Heick Jensen was elected member of the natural science class.
The human genome is promiscuously transcribed yielding RNA from >75% of its DNA, and throughout the years, researchers world-wide have tried to find out how much of this material is functional. Danish researchers have now received a prestigious grant from the Lundbeck Foundation to address this problem.
<em>eLife</em> is a peer-reviewed open access scientific journal for the biomedical and life sciences. It was sponsored by the Howard Hughes Medical Institute, Max Planck Society and Wellcome Trust following a workshop held in 2010 at the Janelia Farm Research Campus and finally established at the end of 2012.
Professor Torben Heick Jensens forskning i genomekspression, stabilitet og teknologi bliver nu støttet af en Advanced Grant fra Det Europæiske Forskningsråd.
Torben Heick Jensen - Professor at the Department of Molecular Biology and Genetics and Director of the Danish National Research Foundation-funded centre for mRNP Biogenesis and Metabolism - has been invited to give a talk in the prestigious ‘Mendel Lecture series’. The event is taking place on 18th April 2013 in Brno, the Czech Republic.
Professor Torben Heick Jensen, Department of Molecular Biology and Genetics, Aarhus University, has achieved great international recognition with his nomination as a member of the European Molecular Biology Organization (EMBO). The election to EMBO is in recognition of Professor Jensen’s outstanding research within gene expression.
The Centre for mRNP Biogenesis and Metabolism studies the crosstalk between transcriptional and post-transcriptional processes, which establishes a network of interdependencies that ultimately regulate gene expression. Our focus is on structure/function relationships of mRNP formation and its quality control as well as the occurrence, and putative function, of non-coding RNA transcription.